Abstract
RNAi has revolutionized genetic research, and is being commercialized as an insect pest control technology. Mechanisms exploited for this purpose are antiviral and therefore rapidly evolving. Ideally, RNAi will also be used for noninsect pests; however, differences in RNAi biology make this uncertain. Tetranychus urticae (two-spotted spider mite) is a destructive noninsect pest, which has a proclivity to develop pesticide resistance. Here we provide a comprehensive study of the endogenous RNAi pathways of spider mites to inform design of exogenous RNAi triggers. This effort revealed unexpected roles for small RNAs and novel genome surveillance pathways. Spider mites have an expanded RNAi machinery relative to insects, encoding RNA dependent RNA polymerase (Rdrp) and extra Piwi-class effectors. Through analyzing T. urticae transcriptome data we explored small RNA biogenesis, and discovered five siRNA loci that appear central to genome surveillance. These RNAs are expressed in the gonad, which we hypothesize to trigger production of piRNAs for control of transposable elements (TEs). This work highlights the need to investigate endogenous RNAi biology as lessons from model organisms may not hold in other species, impacting development of an RNAi strategy.
Keywords: siRNA, piRNA, piRNA cluster, transposons, genome surveillance
INTRODUCTION
RNA interference (RNAi) technology has transformed genetic investigation in model and nonmodel organisms (Kamath and Ahringer 2003; Russell et al. 2017). It was originally described as a process where exogenous double-stranded RNA (dsRNA) could be used to trigger destruction of complementary RNA (Fire et al. 1998). Recently, RNAi is being developed into a promising alternative to broadly toxic pesticides (Joga et al. 2016), and has been successful in controlling several insects (Gordon and Waterhouse 2007; Price and Gatehouse 2008; Huvenne and Smagghe 2010). Yet, RNAi effectiveness can vary dramatically between species suggesting that to be efficient, species-specific design approaches will be needed.
Understanding of RNAi in arthropods is based on nearly two decades of research in Drosophila. In flies, three classes of endogenous small RNAs have been described: microRNAs (miRNAs), short interfering RNAs (siRNAs), and Piwi-interacting RNAs (piRNAs) (Okamura 2012). Each is distinguished by size, biogenesis pathway, and function. miRNAs are deeply conserved across species where siRNAs and piRNAs are not (Wen et al. 2014). siRNAs are primarily an antiviral mechanism in flies, where Dicer2 (Dcr2) processes viral RNAs into 21-nt siRNAs that become trans-acting, destroying viral sequences (Zambon et al. 2006; Gammon and Mello 2015). In flies there are also 21-nt endogenous siRNAs produced from loci that generate dsRNA, such as long inverted repeats or overlapping antisense transcripts (Okamura 2012). siRNA biogenesis in some arthropods is notably influenced by the presence of RNA dependent RNA polymerase (Rdrp) (Lewis et al. 2017). At least three paralogs of Rdrp were present in the most recent common ancestor of Eukaryotes; however, a majority of animal clades lost Rdrp, except nematodes, lophotrochozoans, and chelicerates (spiders, scorpions, horseshoe crabs, ticks, and mites) (Zong et al. 2009). Rdrp has a significant role in Caenorhabditis elegans where it generates amplifying siRNAs and de novo siRNAs (Billi et al. 2014). Rdrp encoding arthropods are thought to generate dsRNAs from single-stranded RNA templates and convert them to Dicer substrates through a mechanism similar to what was found in nematodes (Sarkies et al. 2015; Lewis et al. 2017). In contrast, piRNAs have distinct biogenesis pathways as they are Dicer independent. They are typically associated with germline where they protect integrity of gamete genomes by repressing TEs (Siomi et al. 2011). An important feature of piRNA biology is “master loci” (ML) that catalog restricted sequences (Brennecke et al. 2007). A recent study reported somatic piRNAs in arthropods that target TEs as well as messenger RNAs (Lewis et al. 2017).
Spider mites are chelicerates, diverging from insects and crustaceans nearly 600 million years ago, and is the closest group of arthropods to nematodes from which they diverged approximately 1 billion years ago (Ai et al. 2015). Chelicerates share RNAi features with nematodes such as Rdrp, and have multiple Dicers like insects. How these factors interact is unclear, which has implications for using RNAi to control gene expression in chelicerates. Tetranychus urticae affects agriculture worldwide, and can rapidly develop pesticide resistance, having the maximum incident of pesticide resistance among all arthropods (Grbić et al. 2011; Dermauw et al. 2013). Thus, an additional control method, like RNAi, would be welcomed for mitigating damage caused by these mites. The presence of Rdrp in spider mites suggests that RNAi might be potent as seen in worms. Indeed, there have been reports of trans-generational RNAi silencing in ticks–another chelicerae arthropod that encodes Rdrp (Kocan et al. 2007). However, a recent study reported only modest effectiveness of ingested dsRNA to trigger RNAi in spider mites. (Suzuki et al. 2017). Five methods were tested: feeding on leaf disc floating on dsRNA solutions, rearing animals on transgenic plants expressing dsRNA, feeding dsRNA containing artificial diet, feeding on leaves coated with dsRNA, and soaking mites in dsRNA solution. Only the two most aggressive methods yielded appreciable efficiency; leaf coating and soaking. The challenge of eliciting robust RNAi in spider mites highlighted by this study suggests there may be a gap in the understanding of basic RNAi mechanisms and implementation in this organism.
To understand biogenesis patterns and targets, we used available genome-wide data sets to examine RNAi pathways in a comprehensive way (Grbić et al. 2011). Our analysis shows that spider mites possess an unusual mix of RNAi factors. T. urticae not only has Rdrp but also a more diverse piRNA pathway. We also discovered five siRNA producing loci expressed in the gonad that target transposons and appear to trigger piRNA production. This is the opposite of what is seen in nematodes, which use piRNAs upstream of secondary siRNA production (Girard et al. 2007). Understanding the activity of these derived pathways will be critical for designing potent RNAi in spider mites as it resets expectations for the roles of different small RNA species in this organism's biology. Moreover, our study will benefit efforts to deploy RNAi in other chelicerates as many species in this subphylum possess Rdrp and supernumerary Piwis.
RESULTS AND DISCUSSION
Spider mite RNAi pathways and small RNA producing loci
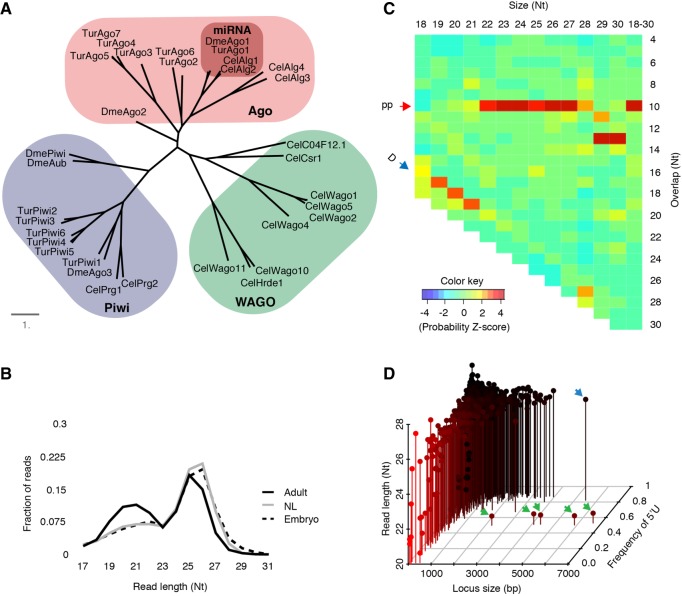
To begin investigation of RNAi pathways in spider mites, we first examined Argonaute/Piwi effector proteins. Distinct Argonautes/Piwis mediate the biology of different classes of small RNAs. Beginning with existing annotations we manually curated thirteen Argonautes/Piwis in T. urticae using genomic and transcriptome data (Supplemental Table 1; Grbić et al. 2011). We then examined their relatedness to Argonautes/Piwis from deer tick, fruit fly, and C. elegans (Fig. 1A). We found that T. urticae Ago1 closely resembles miRNA associated Ago proteins. The remaining six Agos potentially work in siRNA pathways as they clustered with worm Alg-3, Alg-4, and fly Ago2 (Palmer and Jiggins 2015). Presence of six Piwis suggests more elaborate piRNA pathways in T. urticae.
FIGURE 1.
Small RNA effectors and populations in spider mites. (A) Phylogenetic tree showing relationship of Ago proteins from T. urticae (Tur), deer tick- Ixodes scapularis (Isc), Drosophila melanogaster (Dme), and C. elegans (Cel). (B) Size distribution of stage-specific reads mapped to the whole genome. Mapped reads were collapsed using fastx_collapser and the collapsed reads were used to calculate size distribution. (NL) Nymph and larvae. (C) Z-scores for overlap probability of reads that mapped to TE. Red and blue arrow show ping-pong and Dicer cleavage signature, respectively. (pp) Ping-pong; (D) Dicer. (D) Loci were compared by size (x-axis), read length (y-axis), and frequency of “T” residues at the 5′ position (z-axis). Green arrows indicate non-piRNA loci encoded in tandem on scaffold 9. The blue arrow indicates an annotated politron TE.
Next, we analyzed expression patterns of major RNAi factors (Supplemental Fig. 1). Almost half of the annotated Argonaute family members (Ago2,4,5,6/Piwi2,3) showed negligible expression. High expression of Ago7 and Piwi6 was seen in adult animals and Piwi1,4,5 in embryos. Piwi proteins are commonly found to be involved with germline biology, and are functionally coupled with gametogenesis making high expression of Piwis in embryos unexpected. Differential expression of Piwi5 and Piwi6 between embryos and adults suggests that there might be embryo- and adult-specific piRNAs. As somatic piRNAs have been found in arthropods, these embryo-specific Piwis suggest spider mites might also have somatic piRNAs (Lewis et al. 2017). Other RNAi proteins were expressed moderately across stages except for the Rdrps, which were generally low expressed (Supplemental Fig. 1). In spider mites, all Rdrp family members are encoded at a single location on scaffold 2 in the same orientation, perhaps arising from tandem duplication of an ancestral gene (Supplemental Fig. 2). Spider mite Rdrps are predicted to be processive enzymes like C. elegans Rrf-3 (Sarkies et al. 2015), which means they can synthesize long dsRNA using single stranded transcripts as templates and are not involved in generating de novo siRNAs (Supplemental Fig. 3).
Using combined small RNA data sets from mixed gender, whole animal bodies at three developmental stages (adult, nymph-larvae, and embryo) we investigated the composition of spider mite small RNA populations (Supplemental Fig. 4). We observed a bimodal size distribution with peaks at 21–22 and 25–26 nt (Fig. 1B). These two peaks suggest the presence of small RNA classes seen in Drosophila, which is consistent with the repertoire of Ago/Piwi proteins present in the genome of T. urticae. In insects, piRNAs are predominantly involved in TEs suppression, and are produced in two distinct yet collaborative pathways. In flies, a mitochondrial protein Zucchini (Zuc) and Piwi proteins produce “primary” piRNAs in a phasing mechanism from transcripts derived from piRNA clusters, or 3′ UTR regions of protein coding genes, which subsequently trigger production of piRNAs in a so-called ping-pong amplification cycle (Huang et al. 2017). Ping-pong piRNAs overlap by 10 nt due to enzymatic cleavage patterns of Piwi proteins. Ping-pong piRNAs can trigger further Zuc-mediated piRNA synthesis in a self-amplifying system (Han et al. 2015). The T. urticae genome does not appear to encode a zuc ortholog, suggesting a divergent piRNA biology that eschews the Zuc-produced primary-piRNA mechanism.
Next, we investigated if T. urticae small RNAs have a role in genome surveillance, despite the apparent absence of a major biogenesis factor–Zuc. To this end, we identified biogenesis patterns of small RNAs mapping to TE sequences. All mapping events from combined small RNA sequencing data were used to capture all potential RNA–RNA interactions. Alignments were analyzed with an algorithm that can identify overlap probabilities of read pairs in mapping data (Antoniewski 2014). We observed the 10-nt ping-pong signature in TE mapped reads that were longer than 21 nt (Fig. 1C). We also observed the 2 nt overhang Dicer signature in small size TE mapped reads (19–21 nt), which suggests cooccurrence of siRNA and piRNA at TEs. This is consistent with previous observations that small RNAs in the distinct size ranges of siRNAs and piRNAs map to TEs (Grbić et al. 2011).
To characterize the landscape of T. urticae small RNA-producing loci, we called peaks of small RNA expression using uniquely mapping reads. The thousand most highly expressed regions were compared by locus size, average read length, and frequency of 5′ “T” (Fig. 1D; Supplemental Table 2). This analysis recovered many TE loci dispersed throughout the genome, which exhibited a piRNA signature of longer reads and high frequency of 5′ “T.” The largest locus with this profile is a 5.5-kb politron TE. This contrasts with 5 small RNA loci to which substantially shorter reads align that have a significantly lower 5′ “T” bias. These peaks are encoded in close proximity on scaffold 9. Application of this peak calling strategy on a Drosophila whole body, mixed gender small RNA data set recovered major known piRNA clusters (42AB, flam, etc.), but failed to identify loci similar to T. urticae scaffold 9 clusters with the exception of a known siRNA cluster in the flamenco locus (Supplemental Fig. 5; Liu et al. 2011; Guida et al. 2016). The presence of the prominent scaffold 9 loci along with the absence of Zuc reinforces the notion that T. urticae has distinct small RNA-mediated genome surveillance pathways relative to Drosophila.
T. urticae siRNA master loci
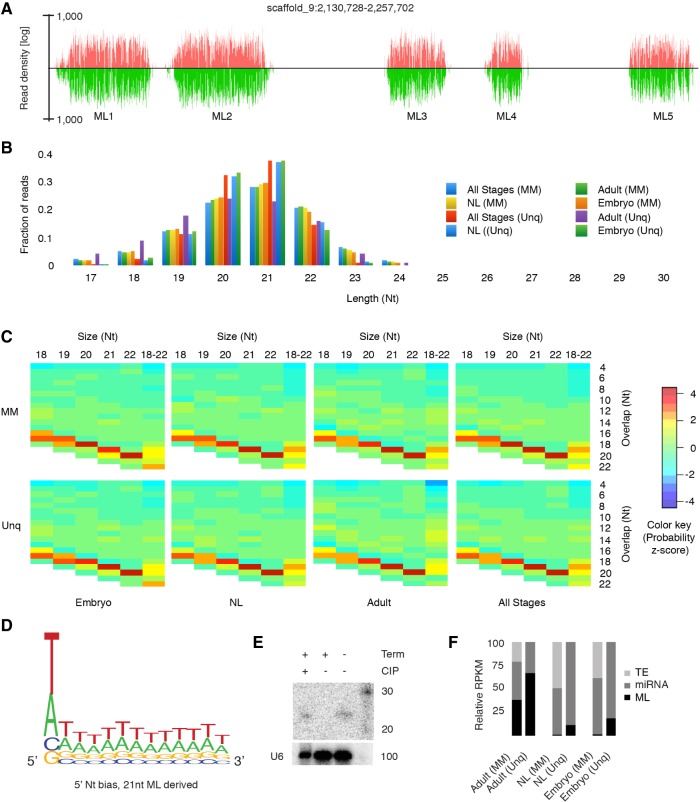
Investigation of RNAi biology in another mite, Dermatophagoides farinae, found a complete loss of piRNA pathways and replacement with Dicer produced, siRNA-mediated genome surveillance, complete with siRNA-based master loci (ML-siRNA) (Mondal et al. 2018). While spider mites clearly have an intact piRNA pathway, they appear to share ML-siRNA loci with dust mites. Indeed, the five small RNA loci on scaffold 9 appear to have many features of ML-siRNA clusters (Fig. 2). While these loci encompass a total length of only ∼30 kb, 16% of reads from the combined sequencing data uniquely map with similar abundance on both strands (Fig. 2A). Almost all the reads were in 19- to 22-nt range with a peak at 21, suggesting they are siRNA-class small RNAs (Fig. 2B). Furthermore, we observed a Dicer-type biogenesis pattern at these loci as overlap Z-scores were highest at −2 lengths (Fig. 2C). This pattern was prominent regardless of developmental stage. We also noticed equal representation of 5′-T/A nucleotide bias at the 5′ position of the reads, and not the high T bias seen in piRNAs (Fig. 2D).
FIGURE 2.
siRNA producing master loci (ML) in spider mite. (A) Uniquely mapped read density of positive (red) and negative (green) at scaffold 9 master loci. (B) Developmental stage-specific read size distribution for both unique (Unq) and all mapping (MM) events. (C) Overhang z-scores of reads produced from the ML from three different developmental stages. (D) 5′-T/A bias of ML mapped reads. (E) Northern blot of ML-siRNA after enzymatic treatment. U6 RNA was used as loading control. (Term) Terminal exonuclease; (CIP) calf-intestinal phosphatase. (F) Relative read density mapping to ML, TE, and miRNA loci (RPKM) in different stages of the spider mite life cycle. (NL) Nymph and larvae.
To gain better insight into biogenesis of the ML-siRNAs, we sought to determine if they have characteristics of Dicer cleavage: 5′-monophosphate and not Rdrp-produced 5′-triphosphates (Lee and Collins 2007). We treated total RNA with the 5′ monophosphate specific terminator ribonuclease. After treatment, complete elimination of ML-siRNAs was observed (Fig. 2E). Terminator-mediated degradation could be abrogated by prior treatment with calf intestinal phosphatase (CIP) (Fig. 2E). This result indicates ML-siRNAs are not generated by a de novo siRNA pathway. We also examined stage-wise relative expression of ML-siRNAs and observed that these loci are primarily expressed in adult compared to other developmental stages indicating they have an adult specific function such as gametogenesis (Fig. 2F). Together, T. urticae ML-siRNAs are Dicer products deriving from a dsRNA precursor, possessing the expected 5′-monophosphate.
ML-siRNAs appear to trigger piRNA production
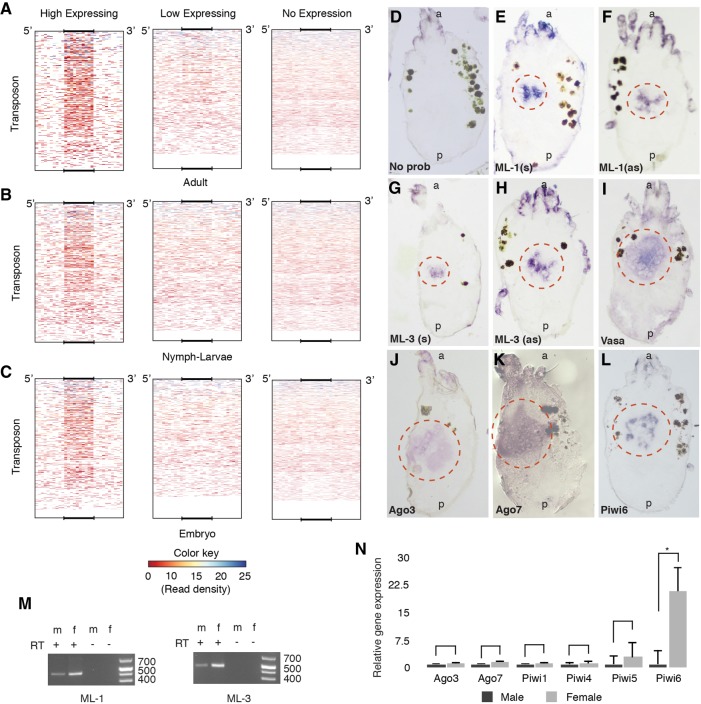
To investigate whether ML-siRNAs have a function like piRNAs produced from piRNA cluster in other animals, we mapped ML uniquely mapping reads back to the whole genome to look for secondary alignments. A mapping strategy was used that captured all mapping events and allowed up to two mismatches per alignment. This approach reveals all potential binding at near perfect complementarity between ML-siRNAs and their targets. This analysis found a significant number of ML-siRNAs can target TEs, and that the frequency of targeting directly correlates to the abundance of piRNAs arising from the TEs (Fig. 3A–C). TE loci were separated into three categories based on piRNA abundance: high, low, and no expression based on density of uniquely mapped, longer reads (24–31 nt). DeepTools was used to count ML-siRNA read depth in TE loci along with 5′ and 3′ flanking regions (Ramírez et al. 2014). From this, we observed a strong correlation between high piRNA abundance at TEs and ML-siRNA mapping (Fig. 3A–C). This was slightly more pronounced in adults compared with other developmental stages (Fig. 3A–C). As the ML-siRNAs only align to scaffold 9 when using unique mapping parameters, but then map to numerous TEs when one to two mismatches are permitted, this allows us to clearly delineate their origin as scaffold 9, and their targets as piRNA processed TE transcripts. This suggests a mechanism where siRNAs trigger piRNAs, which is opposite from the situation in C. elegans where piRNAs trigger siRNAs. This is further corroborated by the absence of Zuc in T. urticae, and implies that siRNAs act like primary piRNAs in this organism. Further dissection of these interactions through genetics will be needed to verify this mechanism.
FIGURE 3.
Master loci siRNAs interact with piRNAs and are expressed in the gonad. Average ML-siRNA read depth in three categories of TE loci in three different developmental stages (A–C). For each stage, TE loci were divided into three groups (1) having >50 longer (24–31) reads, (2) having 1–50 longer reads, and (3) no mapping of longer reads. ML-siRNAs were mapped back to the whole genome, and average read depths were counted using deeptools for each TE group. Mapping to TE coordinates is displayed as a size normalized heat map that includes 500 nt of 5′ and 3′ flanking regions. RNA in situ hybridization following addition of no probe (D), ML-1 (E,F), ML-3 (G,H), Vasa (I), Ago3 (J), Ago7 (K), and Piwi6 (L). (s) Sense strand; (as) antisense strand; (a and p) anterior and posterior of the animals, respectively. Red circles mark gonadal ISH signal. (M) RT-PCR for expression of ML in male and female adult animals. The same loci were amplified in RT-PCR that were used to generate ISH probe. (N) qPCR of ML associated Argonautes and all expressed Piwi proteins, from three independent biological replicates. Error bars represents SEM. Significance was determined by T-test. Asterisk denotes a P-value <0.05.
ML-siRNAs are expressed in the gonad
To better understand the role of ML-siRNAs in spider mite biology, we determined their tissue specificity by in situ hybridization (ISH) in adult female spider mites. Approximately 500-nt sequences from both strands of ML1 and ML3 were used as RNA probes, which revealed female gonad expression (Fig. 3D–I). Gonadal expression was verified by localizing Vasa transcripts—a well-known gonad-specific protein (Fig. 3I; Dearden et al. 2003). A similar signal from hybridization of both sense and antisense probes supports that ML-siRNAs are produced from precursor dsRNAs; canonical substrates of Dicer. To accompany these gonad-specific siRNAs, we also found high expression of ago7 and ago3 in gonads of adult females (Fig. 3J,K).
ML are expressed in both sexes, though higher expression was seen in females (Fig. 3M). We also compared expression of Ago/Piwi in male and female adult mites by RT-qPCR. Piwi6 was significantly more abundant in the female compared to male (Fig. 3N). Together, this suggest that piRNAs, potentially downstream from ML-siRNA expression, may be more active in females. Larger gonad size in females might be the reason for higher female-specific piwi6 expression; however, similar expression levels of piwi1,4,5 and Ago3,7 in both male and female support female-specific expression of piwi6 and that suppressing TEs through collaboration of siRNAs and piRNAs may more be an aspect of oogenesis.
This study provides a thorough analysis of the small RNA biology in T. urticae. In comparison to other arthropods that have been extensively investigated, these chelicerates have distinct RNAi biology. We report the existence of ML-siRNA loci in T. urticae, which produce siRNAs in the gonad and appear similar to loci described in dust mites. Significantly biased mapping of ML-siRNAs to high piRNA targeted-TEs suggests that they might be involved in activation of the ping-pong amplification loop, which is analogous to Zuc-piRNA-mediated triggering of ping-pong (Fig. 4). It is unclear if ping-pong piRNAs feed-forward to promote generation of ML-siRNAs as seen with Zuc-piRNAs, or whether maternally inherited piRNAs contribute to the interaction of piRNAs and siRNAs (Le Thomas et al. 2014). Another outstanding issue is understanding the function of spider mite Rdrp. Is the ML-siRNAs pathway dependent on Rdrp? Does Rdrp activity synergistically interact with ping-pong amplification triggered by ML-siRNAs?
FIGURE 4.
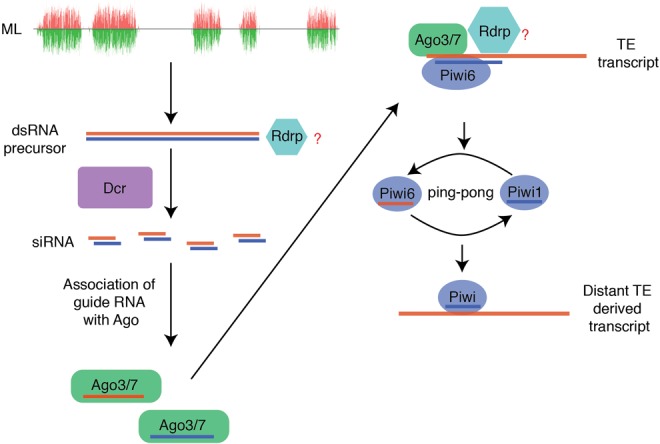
Hypothetical initiation of ping-pong amplification by ML-siRNA in spider mite gonad.
This mechanism does appear to be unique to mites as Ticks possess a Zuc ortholog. Failure to identify a Zuc homolog or large piRNA clusters could be a consequence of incomplete genome assembly, however, this is unlikely due to the small size, low complexity, and method by which the genome was assembled (Grbić et al. 2011). Zuc-processing of piRNAs is conserved between flies and vertebrates, and therefore clearly the ancestral state. Nonetheless the mechanism we described here appears to be effective at controlling TE mobilization as T. urticae has a relatively low TE burden. Furthermore, T. urticae has one of the smallest metazoan genomes, which might have been reduced by rearrangements caused by the loss of Zuc- piRNAs, and the ensuing mobilization of TEs.
Exogenous dsRNA gets incorporated into an antiviral pathway involving Dcr2 and Ago2 in flies. This pathway may be present in spider mites, however, expression of candidate somatic Ago2-like T. urticae genes is negligible (Fig. 1A; Supplemental Fig. 1). Further divergence from flies is evident from embryo-specific Piwi proteins and piRNAs suggests this pathway may not be confined to the germline (Supplemental Fig. 1). How the reconfigured RNAi pathways of spider mites influence the capacity of dsRNA to trigger RNAi is unclear, and highlights the need to investigate metabolism of exogenous dsRNA in spider mites to understand the relative insensitivity reported (Suzuki et al. 2017). Appreciation of spider mite RNAi may also lead to better approaches for controlling other mites like Varroa destructor and citrus mites.
MATERIALS AND METHODS
Argonaute sequence annotation
Amino acid sequences of spider mite Ago proteins were curated from existing annotations using genome and transcriptome data. Transcriptome data was analyzed by Tophat and complete open reading frames (ORFs) of seven Argonaute and six Piwi were verified by manual inspection. ScanProsite was used to detect protein domains (PAZ and PIWI) of the amino acid sequences (de Castro et al. 2006). Ago amino acid sequences of other organisms were downloaded from NCBI. Multiple sequence alignment was accomplished using MUSCLE, phylogeny was constructed using PhyloML and tree was visualized using TreeDyn (Dereeper et al. 2008).
Analysis of the small RNA data sets
Annotations of TE and other genomic elements were downloaded from ORCAE portal (http://bioinformatics.psb.ugent.be/orcae/overview/Tetur). Pipelines used to analyze data sets are shown in Supplemental Figure 4. TE- and ML-specific index files were created using bowtie and the reads were mapped using either all mapping (-a -m 100) options or unique mapping (-v0 -m1 --best --strata) options. Heat map in Figure 1C was generated in R using output from the overlap signature python script (Antoniewski 2014). Nt bias was calculated using seqLogo. Uniquely mapped read depth was determined through bedtools. The thousand highest expressing regions were extracted from read depth data (Supplemental Tables 2, 3). Annotations were established after merging of features within 50 nt.
Counting average read depth at TE loci by deepTools
Longer reads (24–31 nt) were mapped to TEs using bowtie unique mapping options (-v0 -m1 --best --strata), and the number of reads mapped to each locus was counted using bedtools. TE loci were then divided into three bed files: no reads mapped (no expression), 1–50 reads mapped (low expressing), and more than 50 reads mapped per locus (high expressed). ML-siRNAs were mapped to the entire genome using bowtie option -a -m 100 and a bigwig file was created. Bed and bigwig files were used in deepTools through the Galaxy suite to count average read depth per TE locus.
DIG-labeled RNA probe preparation
Approximately 500-nt regions from ML1, ML3, vasa, ago3,7, and piwi6 were amplified by Taq DNA polymerase. PCR product was ligated into the pGEM-T Easy vector (Promega) by T4 DNA ligase. PCR was done using the plasmid as template using primers which both encoded T7 promoter sites. The PCR product was used for in vitro transcription using the MEGAscript T7 Transcription Kit (Thermo Scientific) and DIG RNA labeling mix (Roche). RNA was precipitated using LiCl.
In situ hybridization of adult animal section
Adult female mites were collected and embedded in Tissue-Plus O.C.T compound (Fisher Healthcare). Fourteen-micron sections of whole adult animals were prepared using a cryostat. Sections were dried on glass slides for 20 min at room temperature followed by crosslinking using 4% PFA solution. Crosslinked sections were washed with PBST (PBS plus 0.1% Tween-20) and acetylated for 10 with acetic anhydride (0.25%) in triethanolamine solution. After acetylation, sections were washed in PBST at room temperature and prehybridized at 54°C water bath in hybridization buffer (50% formamide, 4× SSC, 1× Denhardts, 5% dextran sulfate, 250 µg/mL boiled ssDNA, 250 µg/mL tRNA, 50 µg/mL heparin, 0.1% Tween-20) for 2 h. Hybridization of DIG-labeled RNA probe (300 pg/µL) was carried out at 54°C overnight. Following hybridization, sections were washed in wash buffer (50% formamide, 2× SSC, 0.1% Tween-20) for 4 h at 54°C. Sections were incubated in PBST-B solution (PBS, 0.1% Tween, 0.1% BSA) for 30 min at room temperature. AntiDIG-AP (Fab Fragments, Roche) antibody was diluted (1:2000) in PBST-B and sections were incubated in antibody solution for 1.5 h. Sections were washed for 1 h in PBST and incubated in AP buffer (100 mM Tris 9.5, 100 mM NaCl, 50 mM MgCl2, 0.1% Tween-20) for 10 min. Finally, color development was carried out using BM-Purple AP Substrate precipitating solution (Roche) at room temperature.
Northern blot
In each of the reactions, 20 µg of total RNAs were used. In one tube, 1 µL of Terminator exonuclease (epicenter) was added, and exonuclease reaction was carried out for 60 min at 30°C. One microliter of calf intestinal phosphatase (CIP, NEB) was added to a second RNA preparation followed by incubation at 37°C for 30 min. Subsequently, the second preparation was incubated at 30°C for 60 min after adding 1 µL of Terminator exonuclease. RNAs were purified by organic extraction protocol (Goubau et al. 2014). Precipitated RNAs were resolved in urea-polyacrylamide gel (12.5%), and northern blotting was carried out as previously described (Flynt et al. 2009). RNAs were transferred from the gel onto Nylon membrane in 0.5× TBE buffer using 10 V, 300 mA, 1 h at 12°C followed by UV-crosslinking and heating at 80°C for 10 min. Membranes were prehybridized in hybridization buffer (5× SSC, 1 mM EDTA, 2× denhardt's, 1% SDS, 2% dextran sulfate, 30 µg/mL ssDNA) for 1 h at 40°C. Radiolabeling of siRNA oligonucleotide probes was accomplished by incubation with T4 Polynucleotide Kinase (T4 PNK) and P32 gamma-ATP (6000 Ci/mmol). Hybridization was carried out overnight at 40°C followed by washing in 2× SSC, 0.1% SDS for 2 h. Detection of blot signal used phosphorimager screens.
RT-qPCR
One microgram of total RNA from male and female spider mites was used for cDNA synthesis using random hexamer primer. Synthesized cDNAs were used in qPCR assays containing SYBR Green real-time PCR master mix (Thermo Fisher) following manufacturer's protocol.
DATA DEPOSITION
Publicly available sequencing data were used in this study, which can be retrieved under the following accession numbers: GenBank/EMBL/DDBJ (HE587301, HE587940), GEO (GSE32342), and SRA (SRX030911, SRX030913, SRX100283).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eric Riddick and Guadalupe Rojas from the USDA ARS center in Stoneville MS for supplying founders for a Tetranychus urticae Koch colony. The work was supported by NSF-MCB (Award ID: 1616725) and Mississippi INBRE program (P204M103476) from the National Institute of General Medical Science.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.065839.118.
REFERENCES
- Ai C, Zhang Q, Ding J, Ren C, Wang G, Liu X, Tian F, Zhao J, Zhang H, Chen YQ, et al. 2015. Suppression of dust mite allergy by mucosal delivery of a hypoallergenic derivative in a mouse model. Appl Microbiol Biotechnol 99: 4309–4319. [DOI] [PubMed] [Google Scholar]
- Antoniewski C. 2014. Computing siRNA and piRNA overlap signatures. Methods Mol Biol 1173: 135–146. [DOI] [PubMed] [Google Scholar]
- Billi AC, Fischer SE, Kim JK. 2014. Endogenous RNAi pathways in C. elegans. In WormBook (ed. The C. elegans Research Community). WormBook, 10.1895/wormbook.1.170.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. [DOI] [PubMed] [Google Scholar]
- Dearden P, Grbic M, Donly C. 2003. Vasa expression and germ-cell specification in the spider mite Tetranychus urticae. Dev Genes Evol 212: 599–603. [DOI] [PubMed] [Google Scholar]
- de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. 2006. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34: W362–W365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermauw W, Osborne EJ, Clark RM, Grbić M, Tirry L, Van Leeuwen T. 2013. A burst of ABC genes in the genome of the polyphagous spider mite Tetranychus urticae. BMC Genomics 14: 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Flynt A, Liu N, Martin R, Lai EC. 2009. Dicing of viral replication intermediates during silencing of latent Drosophila viruses. Proc Natl Acad Sci 106: 5270–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammon DB, Mello CC. 2015. RNA interference-mediated antiviral defense in insects. Curr Opin Insect Sci 8: 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard LR, Fiedler TJ, Harris TW, Carvalho F, Antoshechkin I, Han M, Sternberg PW, Stein LD, Chalfie M. 2007. WormBook: the online review of Caenorhabditis elegans biology. Nucleic Acids Res 35: D472–D475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon KH, Waterhouse PM. 2007. RNAi for insect-proof plants. Nat Biotechnol 25: 1231–1232. [DOI] [PubMed] [Google Scholar]
- Goubau D, Schlee M, Deddouche S, Pruijssers AJ, Zillinger T, Goldeck M, Schuberth C, Van der Veen AG, Fujimura T, Rehwinkel J, et al. 2014. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature 514: 372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbić M, Van Leeuwen T, Clark RM, Rombauts S, Rouzé P, Grbić V, Osborne EJ, Dermauw W, Ngoc PC, Ortego F, et al. 2011. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 479: 487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guida V, Cernilogar FM, Filograna A, De Gregorio R, Ishizu H, Siomi MC, Schotta G, Bellenchi GC, Andrenacci D. 2016. Production of small noncoding RNAs from the flamenco locus is regulated by the gypsy retrotransposon of Drosophila melanogaster. Genetics 2014: 631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BW, Wang W, Li C, Weng Z, Zamore PD. 2015. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348: 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Fejes Tóth K, Aravin AA. 2017. piRNA biogenesis in Drosophila melanogaster. Trends Genet 33: 882–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huvenne H, Smagghe G. 2010. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J Insect Physiol 56: 227–235. [DOI] [PubMed] [Google Scholar]
- Joga MR, Zotti MJ, Smagghe G, Christiaens O. 2016. RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: what we know so far. Front Physiol 7: 553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321. [DOI] [PubMed] [Google Scholar]
- Kocan KM, Manzano-Roman R, de la Fuente J. 2007. Transovarial silencing of the subolesin gene in three-host ixodid tick species after injection of replete females with subolesin dsRNA. Parasitol Res 100: 1411–1415. [DOI] [PubMed] [Google Scholar]
- Lee SR, Collins K. 2007. Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat Struct Mol Biol 14: 604–610. [DOI] [PubMed] [Google Scholar]
- Le Thomas A, Stuwe E, Li S, Du J, Marinov G, Rozhkov N, Chen YC, Luo Y, Sachidanandam R, Toth KF, et al. 2014. Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev 28: 1667–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SH, Quarles KA, Yang Y, Tanguy M, Frézal L, Smith SA, Sharma PP, Cordaux R, Gilbert C, Giraud I, et al. 2017. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat Ecol Evol 2: 174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Abe M, Sabin LR, Hendriks GJ, Naqvi AS, Yu Z, Cherry S, Bonini NM. 2011. The exoribonuclease Nibbler controls 3′ end processing of microRNAs in Drosophila. Curr Biol 21: 1888–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal M, Klimov P, Flynt AS. 2018. Rewired RNAi-mediated genome surveillance in house dust mites. PLoS Genet 14: e1007183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K. 2012. Diversity of animal small RNA pathways and their biological utility. Wiley Interdiscip Rev RNA 3: 351–368. [DOI] [PubMed] [Google Scholar]
- Palmer WJ, Jiggins FM. 2015. Comparative genomics reveals the origins and diversity of arthropod immune systems. Mol Biol Evol 32: 2111–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DR, Gatehouse JA. 2008. RNAi-mediated crop protection against insects. Trends Biotechnol 26: 393–400. [DOI] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. 2014. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42: W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JJ, Theriot JA, Sood P, Marshall WF, Landweber LF, Fritz-Laylin L, Polka JK, Oliferenko S, Gerbich T, Gladfelter A, et al. 2017. Non-model model organisms. BMC Biol 15: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkies P, Selkirk ME, Jones JT, Blok V, Boothby T, Goldstein B, Hanelt B, Ardila-Garcia A, Fast NM, Schiffer PM, et al. 2015. Ancient and novel small RNA pathways compensate for the loss of piRNAs in multiple independent nematode lineages. PLoS Biol 13: e1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 12: 246–258. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Nunes MA, España MU, Namin HH, Jin P, Bensoussan N, Zhurov V, Rahman T, De Clercq R, Hilson P, et al. 2017. RNAi-based reverse genetics in the chelicerate model Tetranychus urticae: a comparative analysis of five methods for gene silencing. PLoS One 12: e0180654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Mohammed J, Bortolamiol-Becet D, Tsai H, Robine N, Westholm JO, Ladewig E, Dai Q, Okamura K, Flynt AS, et al. 2014. Diversity of miRNAs, siRNAs, and piRNAs across 25 Drosophila cell lines. Genome Res 24: 1236–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon RA, Vakharia VN, Wu LP. 2006. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell Microbiol 8: 880–889. [DOI] [PubMed] [Google Scholar]
- Zong J, Yao X, Yin J, Zhang D, Ma H. 2009. Evolution of the RNA-dependent RNA polymerase (RdRP) genes: duplications and possible losses before and after the divergence of major eukaryotic groups. Gene 447: 29–39. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.