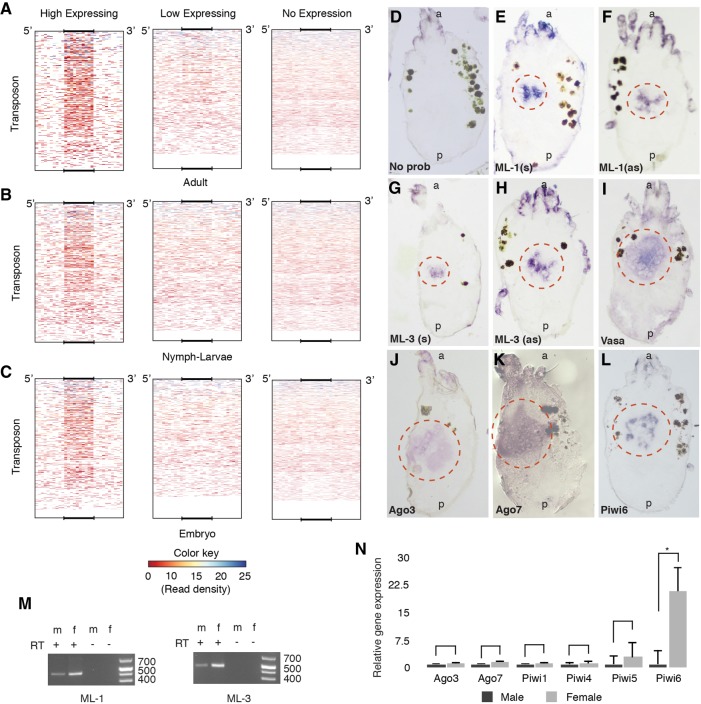
FIGURE 3.
Master loci siRNAs interact with piRNAs and are expressed in the gonad. Average ML-siRNA read depth in three categories of TE loci in three different developmental stages (A–C). For each stage, TE loci were divided into three groups (1) having >50 longer (24–31) reads, (2) having 1–50 longer reads, and (3) no mapping of longer reads. ML-siRNAs were mapped back to the whole genome, and average read depths were counted using deeptools for each TE group. Mapping to TE coordinates is displayed as a size normalized heat map that includes 500 nt of 5′ and 3′ flanking regions. RNA in situ hybridization following addition of no probe (D), ML-1 (E,F), ML-3 (G,H), Vasa (I), Ago3 (J), Ago7 (K), and Piwi6 (L). (s) Sense strand; (as) antisense strand; (a and p) anterior and posterior of the animals, respectively. Red circles mark gonadal ISH signal. (M) RT-PCR for expression of ML in male and female adult animals. The same loci were amplified in RT-PCR that were used to generate ISH probe. (N) qPCR of ML associated Argonautes and all expressed Piwi proteins, from three independent biological replicates. Error bars represents SEM. Significance was determined by T-test. Asterisk denotes a P-value <0.05.