Figure 6.
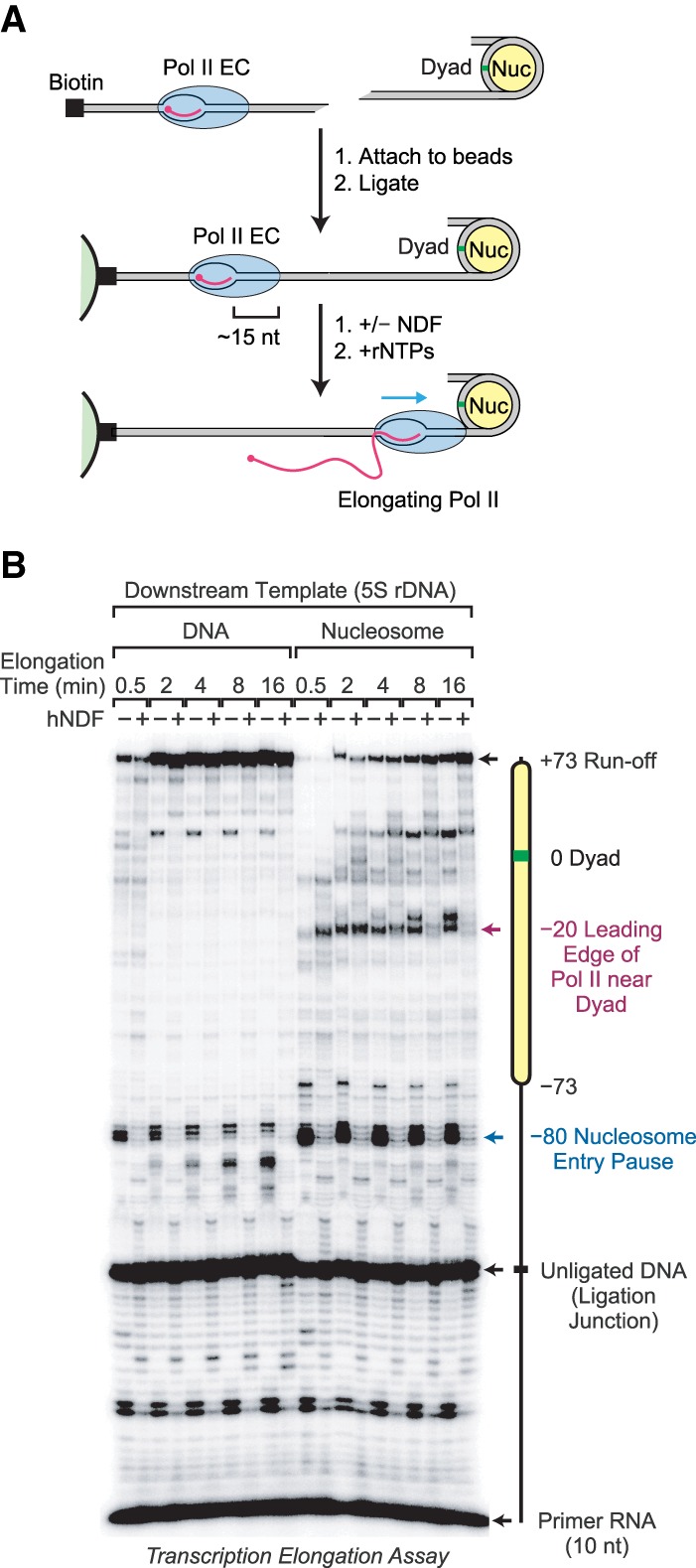
NDF facilitates transcription elongation through a nucleosome. (A) Schematic diagram of Pol II transcription through a positioned nucleosome. A transcriptionally engaged elongation complex (Pol II EC) was assembled with purified yeast Pol II and a 5′ end-labeled 10-nucleotide (nt) RNA primer. This complex was attached to streptavidin beads and then ligated to a downstream mononucleosome positioned on the 5S rDNA sequence from Xenopus borealis. Transcription elongation was initiated by the addition of ribonucleoside 5′-triphosphates (rNTPs). Where indicated, NDF was added after the ligation step but before the addition of the rNTPs. In parallel experiments, naked DNA (same 5S rDNA sequence) was ligated downstream from the elongation complex instead of a mononucleosome. The distance (∼15 nt) from the leading edge of Pol II to the 3′ end of the transcript is also shown. (B) NDF reduces the inhibition of Pol II elongation by a nucleosome. Transcription elongation reactions were performed as described in A in the presence or absence of purified hNDF for the indicated times. Experiments with nucleosomal templates included a 75-fold molar excess (0.3 µM) of free unligated mononucleosomes, which stabilize the low concentration of immobilized nucleosomes. Where indicated, hNDF was included at a concentration of 1.5 µM. The reaction products were resolved by 8% polyacrylamide–urea gel electrophoresis. The diagram shows the locations of the positioned nucleosomes, the sites of Pol II pausing, the runoff product, the ligation junction, and the 10-nt primer RNA. The sizes of the RNA species were estimated by comparison with a 25-nt radiolabeled DNA ladder.
