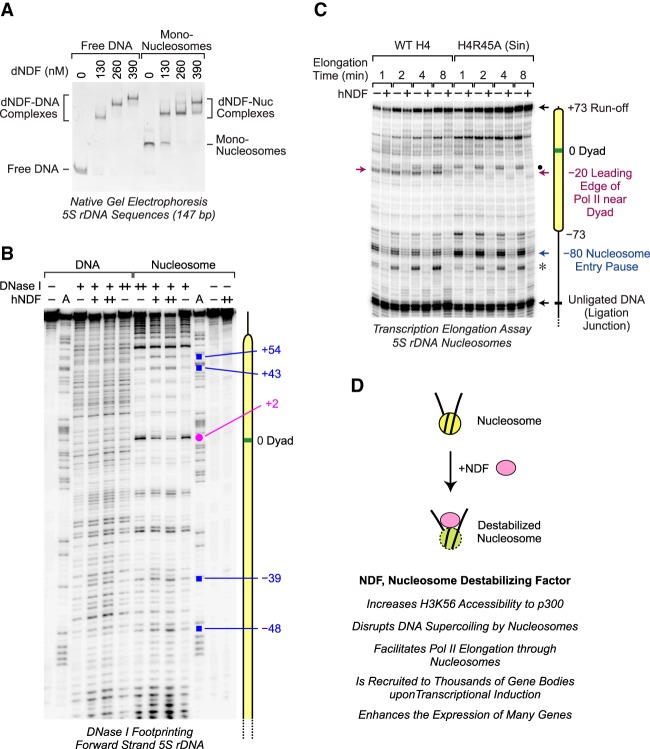
Figure 7.
NDF interacts with nucleosomes near the dyad. (A) NDF binds to mononucleosomes as well as to free DNA. Gel mobility shift assays were performed with the indicated concentrations of dNDF and 50 nM 5S rDNA (147 bp) as either naked DNA or mononucleosomes. The resulting species were separated by 4% nondenaturing polyacrylamide gel electrophoresis and visualized by staining with ethidium bromide. The positions of the shifted complexes are indicated. (B) DNase I footprinting analysis reveals an interaction of NDF near the nucleosome dyad. DNase I footprinting reactions were carried out with the forward strand of the 5S rDNA sequence (183 bp) as either naked DNA or mononucleosomes. Purified hNDF (73 or 150 nM concentration) was included where indicated. The Maxam-Gilbert A sequencing ladder was used to map the locations of the DNase I digestion products. The pink dot shows an NDF-protected nucleotide at +2 relative to the nucleosomal dyad at position 0. The blue squares designate bands that are enhanced in the presence of hNDF. The forward strand has the same sense as the template strand of the 5S rRNA gene. (C) Comparison of the effects of NDF and the H4R45A Sin mutation on Pol II elongation through a nucleosome. Transcription elongation experiments were performed as in Figure 6 with either wild-type histone H4 or mutant histone H4R45A. The red arrow denotes the −20 Pol II pause site that is lost upon mutation of H4R45. The black dot shows the location of a pause site downstream from the −20 position that accumulates with H4R45A nucleosomes. The asterisk corresponds to a pause site that likely derives from the presence of some downstream naked DNA, as in the left panel of Figure 6B. (D) Model for the function of NDF.