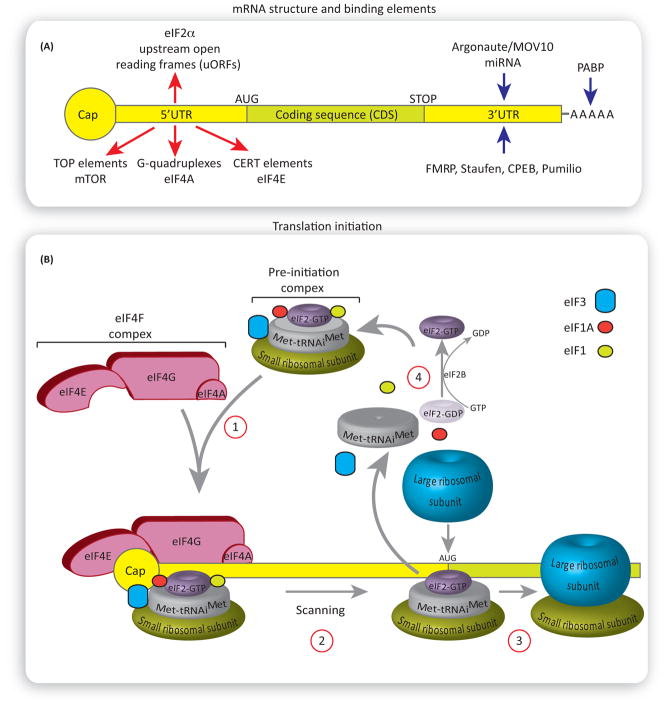
Figure 1. Regulation of Translation Initiation.
(A) General structure of the mammalian mRNA and its binding elements. Red arrows show the 5′ untranslated region (UTR) elements rendering selective translation of specific mRNAs. mRNAs harboring G-quadruplexes in their 5′UTR are dependent on the activity of eukaryotic initiation factor 4F (eIF4A) helicase for their translation. Cytosine-enriched regulator of translation (CERT) renders eIF4E selectivity. Translation of mRNAs with 5′ terminal oligopyrimidine tract (TOP) is mammalian of rapamycin complex 1 (mTORC1) sensitive. Blue arrows show binding elements that regulate the rates of mRNA translation via binding the 3′UTR or poly(A) tail. (B) An overview of translation initiation. The eIF4F complex recruits the pre-initiation complex to the mRNA (1). The pre-initiation complex scans the 3′UTR in a 5′ to 3′ direction for the presence of the start codon, AUG. Once it encounters the start codon, the eIF2-GDP and other factors (eIF1, eIF3, eIF4F, eIF4B, and eIF5) are displaced, followed by binding of the large ribosomal subunit 60S (2) to form the 80S ribosomal initiation complex, which proceeds to the elongation step (3). eIF2B catalyzes the recycling of GDP-eIF2 to GTP-eIF2, allowing for a new round of translation initiation (4).