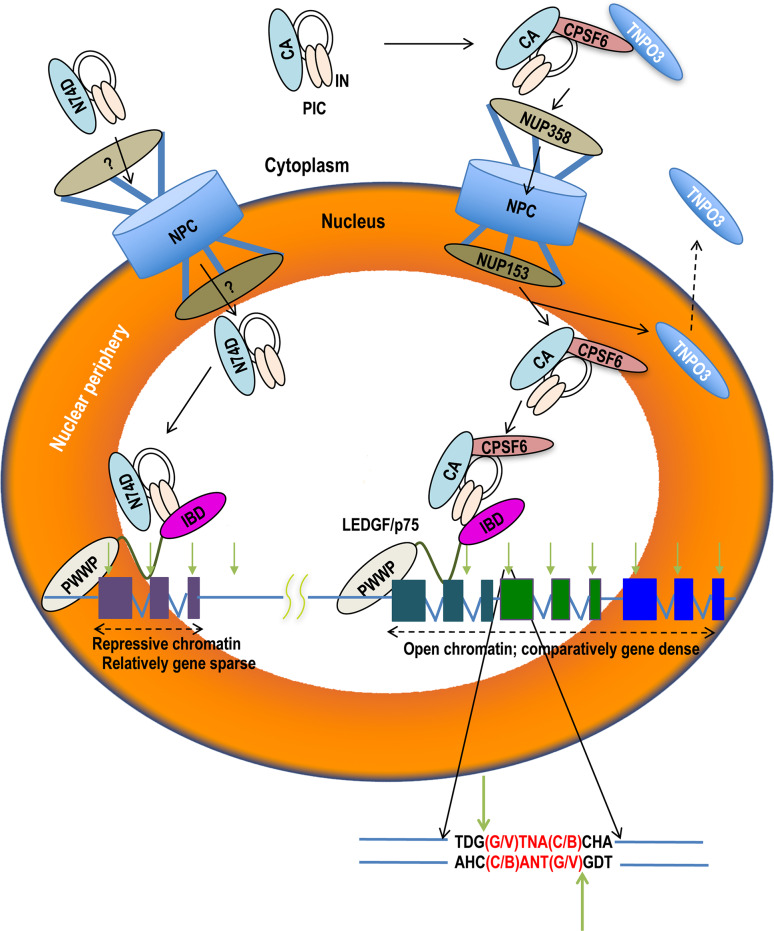
Fig. 4.
Cellular and molecular mechanisms of HIV-1 integration targeting. The HIV-1 PIC harbors viral DNA (black lines), integrase (IN, light orange ovals), and capsid (CA). CPSF6 may bind CA in the cytoplasm to facilitate PIC nuclear import in a NUP358 and NUP153-dependent manner; CPSF6 is shown in association with transportin 3 (TNPO3), one of its known β-karyopherin-binding partners. Following nuclear import, TNPO3 will recycle to engage additional transport substrates in the cytoplasm. The PIC utilizes both CPSF6 and LEDGF/p75 to target integration to active genes (represented as three colored exons separated by introns) preferentially located within the peripheral region of the nucleus (orange shade). A typical integration site (downward light green arrow) is enlarged below to indicate nucleotide sequence preference in International Union of Biochemistry base code (B: G, C, or T; D: G, A, or T; H: C, A, or T; N: G, C, A, or T; V: G, C, or A); opposing green arrows denote scissile phosphodiester bonds. PICs that cannot properly engage CPSF6, as represented by the N74D change in CA, enter the nucleus via an alternate route (marked ?) that may require NUP155 [27]. Such PICs prefer gene sparse regions and hyper target the peripheral region of the nucleus for integration [25, 129, 136, 142, 152]