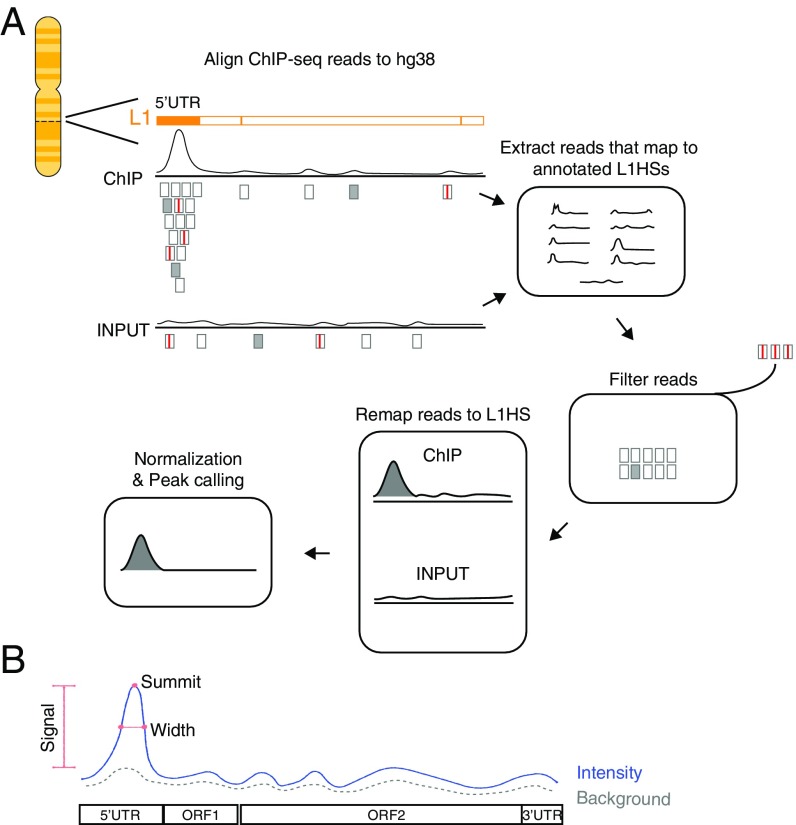
Fig. 1.
The pipeline of MapRRCon. (A) In this pipeline, ChIP-seq data are first aligned to the human reference sequence hg38. Both unique reads (filled gray boxes) and multiply aligned reads (hollow gray boxes) are then extracted and mapped to the 1,620 annotated L1HS sites based on their genome coordinates. We exclude reads with partial alignment (soft clipping), more than three mismatches, or any indels (boxes with red lines). Filtered reads are subsequently mapped to the L1HS consensus sequence to obtain compiled reads. Finally, we generate coverage profiles for both ChIP and Input data and then perform median-based normalization (Methods). The normalized data are used for peak calling. (B) We developed a peak-calling algorithm that is suitable for short sequences such as L1s. Peaks are detected by applying a smoothing filter and finding positions where the smoothed signal has maxima. The peaks are filtered using two thresholds on the original signal: signal intensity (blue line) minus background intensity (dotted gray line) larger than 1 and an rmsd ratio between signal and background larger than 1.3. The width of the peak (red line) is defined by the location where the signal drops to 25% of its maximum.