Figure 1.
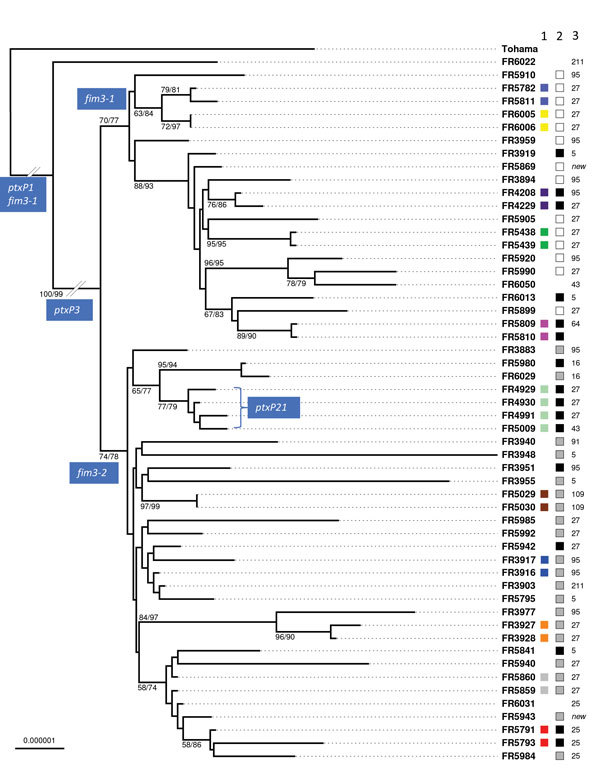
Maximum-likelihood phylogenetic tree for Bordetella pertussis isolates based on the concatenated multiple sequence alignments of 2,038 core genome multilocus sequence typing loci. The tree was rooted on the Tohama reference isolate (GenBank accession no. NC_002929). Only branch support values >50 are labeled (bootstrap/aLRT-SH). Column 1 to the right of the isolates’ names shows colors indicating the intrafamilial groups or groups of multiple isolates from single patients (corresponding to colors shown in inner circle of Figure 2). Column 2 shows pulsed-field gel electrophoresis types (French nomenclature; white, IVα; gray, IVβ; black, IVγ). Column 3 shows multilocus variable-number tandem-repeat analysis types. Scale bar indicates nucleotide substitutions per site.
