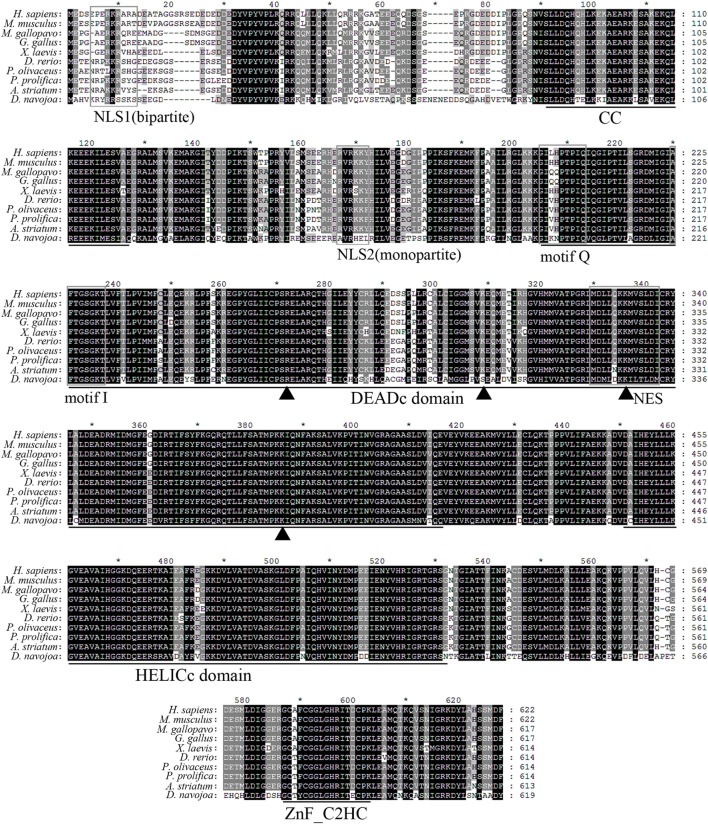
Figure 1.
Multiple alignment of Danio rerio DDX41 with other orthologs. Residues shaded in black are completely conserved across all the species aligned, whereas residues shaded in gray are similar with respect to side chains. The dashes in the amino acid sequences indicate gaps introduced to maximize alignment. The four conserved domains coiled-coil (CC), DEADc, HELICc, and ZnF_C2HC are indicated below the alignment. The nuclear localization signal (NLS)1, NLS2, motif Q, motif I, and nuclear export signal (NES) consensus sequences are boxed. Triangles indicate the four important residues in the double-stranded DNA or cyclic dinucleotide binding.