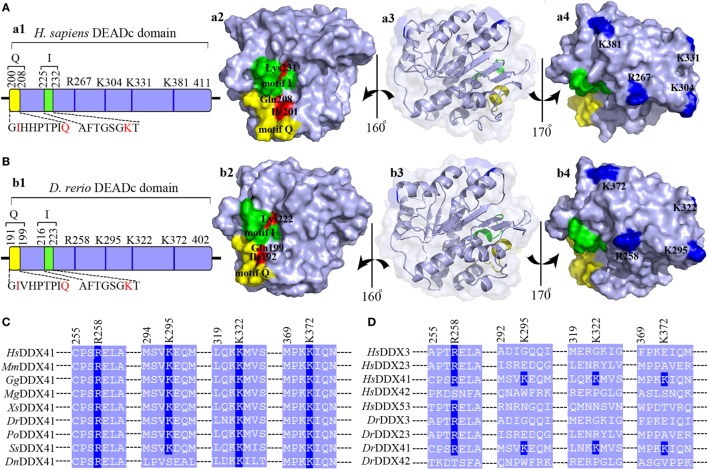
Figure 2.
Motif organization of the DEADc domain. Schematic diagrams and tertiary structures of the functional motifs in DEADc domains of humans [(A), in an APO open form; PDB ID: 5GVS] and zebrafish (B) were presented. (a1,b1) DEADc domain of humans (a1) and zebrafish (b1) contains a motif Q (yellow) and a motif I (green) at its N-terminal region and the key arginine (R) and lysine (K) residues (deep blue) at its C-terminal region (light blue). The color codes representing the motifs and the N- or C-terminal regions are used similarly in the other figures, unless otherwise stated. (a2,b2) Molecular surface representation of the residues involving in recognizing the adenine moiety and the β- and γ-phosphate groups of ATP in Homo sapiens DDX41 (HsDDX41) (a2) and Danio rerio DDX41 (DrDDX41) (b2) in motif Q and motif I. (a3,b3) Molecular surface and α-helixes and β-sheets representation of DEADc domain in a same view with Figures S4C,D (lower). (a4,b4) Molecular surface representation of the residues involving in the putative double-stranded DNA (dsDNA)/cyclic dinucleotide (CDN) binding (colored in deep blue). (C) Amino acid sequence of the residues near the putative dsDNA/CDN-binding sites of DrDDX41 and its orthologs (C), as well as its paralogs (D). Residues critical for dsDNA or CDN binding are indicated in deep blue. The accession numbers for the sequences of DDX41 and its orthologs and paralogs are shown in Figure S3.