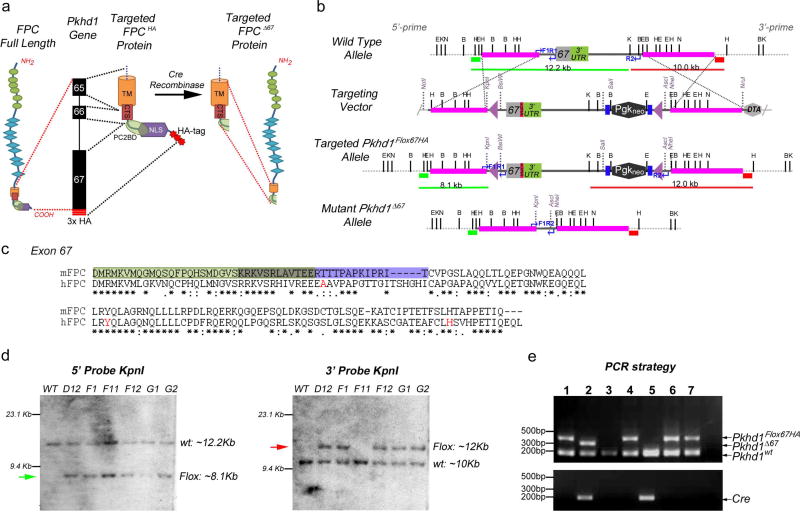
Figure 1. Gene-targeting strategy used to generate a floxed HA-tagged allele for murine Pkhd1 (Pkhd1Flox67HA).
(a) Fibrocystin/Polyductin (FPC) Protein Targeting Strategy. The location of C-terminal protein domains within Pkhd1 exons 65–67 are indicated. The targeted FPC protein includes three HA-tags (red bars or stars) at its C-terminus. Cre recombinase mediated deletion results in loss of the targeted intracellular domain (ICD) containing the nuclear localization signal (NLS), the polycystin 2 binding domain (PC2BD) and the HA tags. The transmembrane domain (TM) and the ciliary targeting sequence (CTS) remain intact. (b) Pkhd1 Gene Targeting Strategy. Genomic maps of the wild-type Pkhd1 allele, the targeting vector, the Pkhd1Flox67HA allele after homologous recombination and following Cre-mediated deletion (Pkhd1Δ67) are indicated. Three HA-tags (red bar) were inserted in frame after exon 67 and before the stop codon. A neomycin cassette flanked by two FRT sites (blue bars) was cloned adjacent to the 3′ UTR and LoxP sites (purple triangles) were inserted into intron 66 and outside of the 3′ UTR. Neomycin was used for positive selection and DTA for negative selection of ES cells. The pink bars represent homology arms while the green and red bars indicate the 5’ and 3’ probes, respectively, that were used for Southern blot analysis. The blue arrows indicate the PCR primers developed for genotyping. Single letters identify the following restriction sites: B, BglII; E, EcoRV; H, HindIII; X, XbaI, K, Kpnl and N, Notl. (c) Alignment of murine FPC exon 67 (137 amino acids) with the homologous region of human FPC. The light green box indicates the putative PC2 binding domain that overlaps (olive box) with the nuclear localization signal (purple box). The amino acid residues in red are affected by pathogenic mutations in ARPKD patients. Asterisks indicate conserved residues. (d) Autoradiographs of Southern blots prepared from targeted ES cell clones (D12, F1, F11, F12, G1 and G2) digested with KpnI. Blots were hybridized to 5’ (green bar) and 3’ (red bar) probes in Panel B. Fragment sizes that correspond to those predicted for the wild type and targeted or floxed (green and red arrows) allele are shown. Clones D12, F1, F12, G1 and G2 yielded fragments of the correct sizes using both probes. (e) Three-primer PCR genotyping strategy that distinguishes 3 alleles Pkhd1wt, Pkhd1Flox67HA and Pkhd1Δ67. The location of primers and the size of expected bands are indicated in Panel b (Pkhd1wt 188bp, Pkhd1Flox67HA 320bp and Pkhd1Δ67 268bp). Cre recombinase specific PCR is shown in the bottom panel. Pkhd1Flox67HA heterozygotes were bred with Meox2tm1(Cre)Sor/J mice to delete Pkhd1 exon 67. Four animals (Lanes 1, 4, 6 and 7) were Pkhd1Flox67HA/WT compound heterozygotes. The Pkhd1Δ67 band is not seen since the animals are Cre negative. Two animals (Lanes 3 and 5) were Pkhd1wt homozygotes. The animal in lane 2, which is Cre positive, has the genotype Pkhd1Δ67/WT after deletion of exon 67.