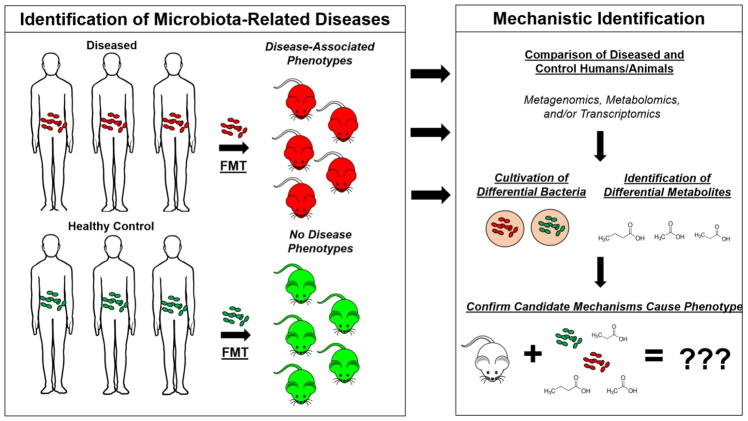
FIGURE 1. From microbiome discovery to mechanism.
An example of the path from observing the microbiome may be involved in a disease to a mechanistic understanding. One approach to explore whether or not the microbiota is involved in a given disease is to transfer the gut microbiota from a patient suffering a disease into an animal via fecal microbiome transplant (FMT) and then pass that animal through the appropriate disease model. If transplantation of the gut microbiota from a diseased patient affects the end points in the model (but transplant of a microbiota from health controls do not), effort should go into understanding a potential underlying mechanism. Generally, this is achieved by using a broad -omic approaches, ideally through the combination of metagenomics, metabolomics, and/or transcriptomics of host stool and other tissues. By comparing the results from disease-presenting animals to controls, candidate bacterium and/or metabolites that may be influencing the disease end points can be identified. If introduction of the candidate trigger organism(s) or metabolite(s) results in the same change in end points, it is likely they are involved in presentation of the phenotypes.