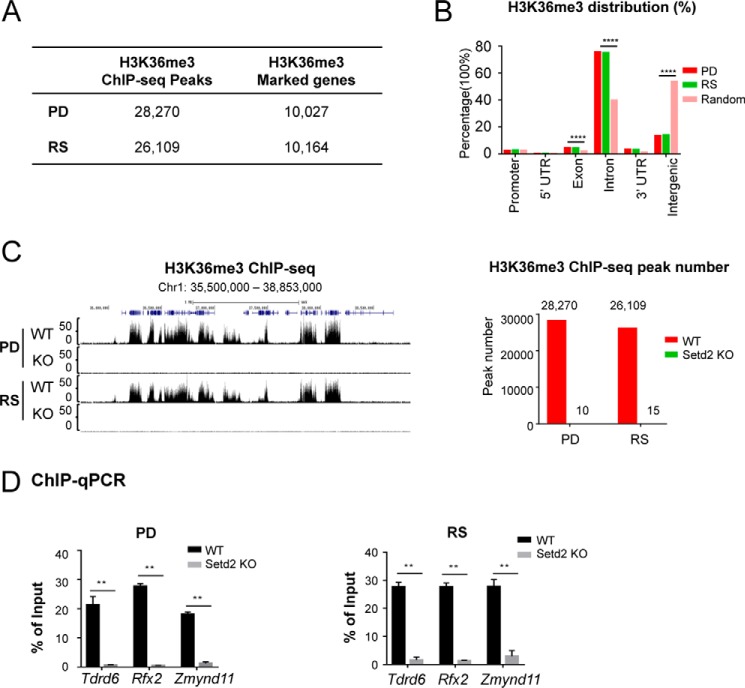
Figure 5.
Setd2 inactivation led to global loss of H3K36me3 in pachytene spermatocytes and round spermatids. A, table summarizing the numbers of H3K36me3 ChIP-seq peaks and H3K36me3-covered genes in PD and RS cells. B, genomic distribution of H3K36me3 ChIP-seq peaks in PD and RS cells compared with genome background. ****, p < 0.0001, one-sided binomial test. C, global loss of H3K36me3 in both PD and RS cells from Setd2 cKO mice. Left, UCSC Genome Browser track showing H3K36me3 ChIP-seq signals at a select genomic region in control and Setd2 cKO mice. Right, numbers of H3K36me3 ChIP-seq peaks in PD and RS cells from control and Setd2 cKO. D, global loss of H3K36me3 in Setd2 cKO mice was confirmed by ChIP-qPCR. q-PCR data are represented as mean ± S.D. from two biological replicates. **, p < 0.01, t test. Error bars represent S.D.