Figure 6.
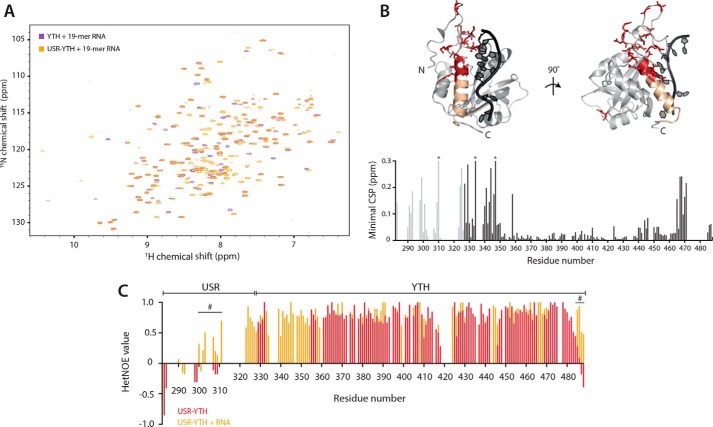
The USR influences the RNA-bound N- and C-clamps. 2D-NMR spectral analysis of YTH and USR–YTH constructs bound to a 19-mer DSR-containing RNA. A, overlay of 1H-15N BEST-TROSY spectra. B, nearest neighbor CSP maps (bottom). Lighter gray peaks denote residues present in the USR–YTH construct but not the YTH construct. Asterisks mark chemical shifts >0.3 ppm. Regions showing large chemical shift differences (above 0.1 ppm; red) or line broadening (yellow) are mapped onto the crystal structure of the Mmi1 YTH domain (top). C, plot of 1H-15N heteronuclear NOE values per residue in the absence (red) and presence (orange) of RNA. Positive values approaching 1 represent more ordered amide backbone regions. Peak absences represent residues that are not assigned. Many N-terminal residues that appear on RNA binding have hetNOE values that suggest they are ordered. The hash symbol marks some regions, assigned in both the absence and presence of RNA, with enhanced rigidity in the presence of RNA.