Figure 8.
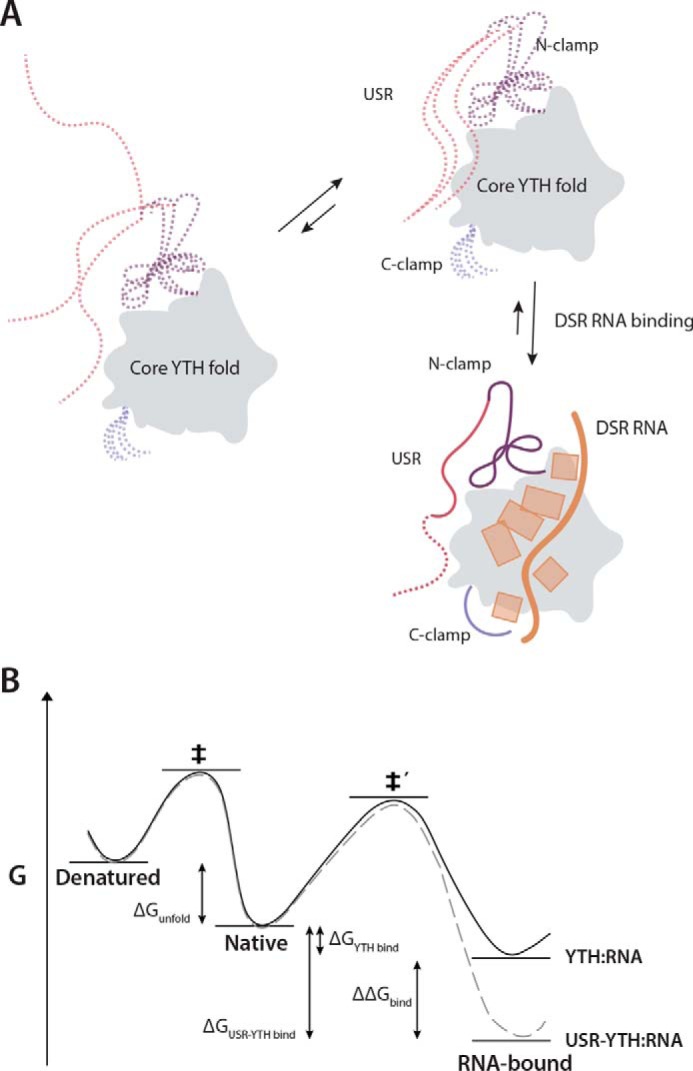
Model for RNA binding by Mmi1. A, schematic diagram showing the contribution of different regions of Mmi1 to DSR RNA binding. The core, conserved YTH fold acts as a platform for RNA binding but is unable to bind RNA with high affinity alone. The USR, N-clamp, and C-clamps have no secondary structure and are dynamic in the absence of RNA (dashed lines). The USR and N-clamp likely exist in an equilibrium between multiple conformers, at least one of which has high affinity for RNA. On RNA binding, these regions become more ordered and contact or reinforce contacts with RNA. RNA backbone and bases are shown in orange. B, thermodynamic model of USR–YTH interaction with RNA. The presence of the USR (gray dashed line) does not affect the thermodynamic stability of the native protein compared with YTH alone (black line). Hence, as there is no change in the energy for unfolding (ΔGunfold) to the denatured state, the apparent melting temperatures are the same. The USR lowers the energy of the RNA-bound complex (ΔΔGbind) via changes in conformational dynamics and possibly a transient interaction with the RNA. Consequently, although the energy barrier for binding (native to ‡′) will be unaffected and similar association kinetics are observed with and without USR, the barrier for dissociation has increased, leading to significantly slower dissociation rates for USR–YTH/RNA compared with YTH/RNA.
