Figure 3.
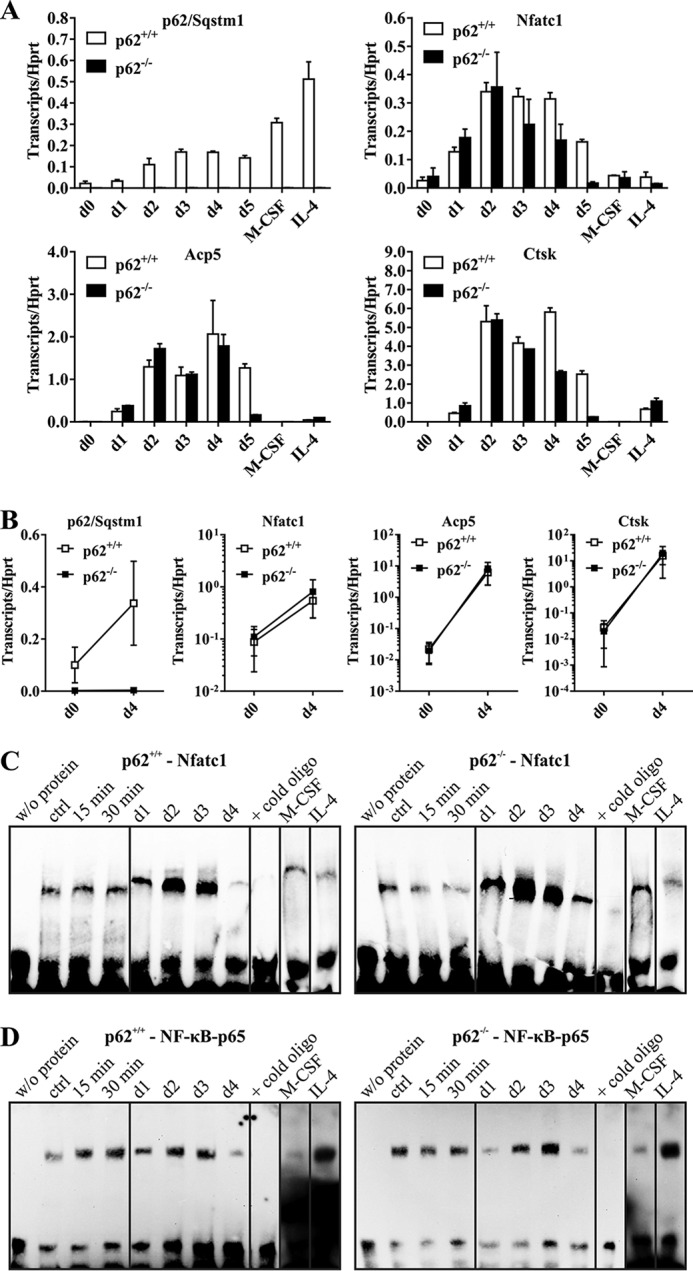
A, p62, Nfatc1, Acp5, and Ctsk gene expression in p62+/+- and p62−/−-derived cells (C57BL/6N) during OCG. cDNAs were obtained from p62+/+- and p62−/−-derived RNA samples isolated from cells at the indicated OCG time points. p62 KO cells produce a significant induction of Nfatc1, Acp5, and Ctsk during OCG despite the verified p62 deficiency. Hprt was used for normalization. Data are mean ± S.D. (error bars) of duplicates. B, gene expression data of at least three independent OCG approaches at days (d) 0 and 4. C and D, EMSA-based verification of Nfatc1 and NF-κB p65 activation during OCG with either p62+/+- or p62−/−-derived OC progenitors (C57BL/6N). BMMs were stimulated with M-CSF/sRANKL for 15 or 30 min (short time) or 1–4 days (long time). Nuclear extracts were analyzed for Nfatc1 (C) and NF-κB p65 (D) activation. The validity of the experiments was confirmed by indicated controls (w/o protein, excess of nonbiotinylated oligonucleotides, M-CSF only, IL-4–mediated inhibition of OCG; ctrl, control). WT and p62−/−-derived OCs show activation of Nfatc1 and NF-κB p65 at days 2 and 3 of M-CSF/sRANKL–induced OCG.
