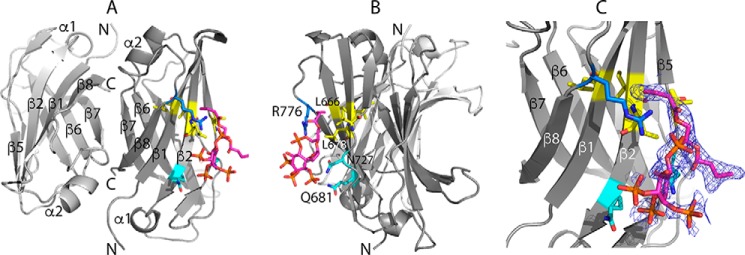
Figure 5.
X-ray diffraction structure of the C2(C771A)–PI(3,4,5)P3 complex. Shown are three views of the complex (Protein Data Bank code 6FJC) in which rotation of view A about the vertical axis through ∼90° gives view B. PI(3,4,5)P3 binds to chain A of an antiparallel C2(C771A) dimer as does PI(4,5)P2. C, a closer view of the C2(C771A)–PI(3,4,5)P3 interaction in the same orientation as A with electron density obtained in a simulated annealing omit map around the PI(3,4,5)P3-binding site shown in blue mesh, contoured at 1.1σ. PI(3,4,5)P3 is shown mainly in magenta. Some of the C2 residues that interact with PI(3,4,5)P3 are shown (hydrophobic residues in yellow, polar residues in cyan, and positively charged residues in blue), including the side chains of Leu-678, Gln-681, Asn-727, and Arg-776 and backbone of Leu-666 and Ile-677. Oxygen and nitrogen atoms are shown in red and dark blue, respectively.