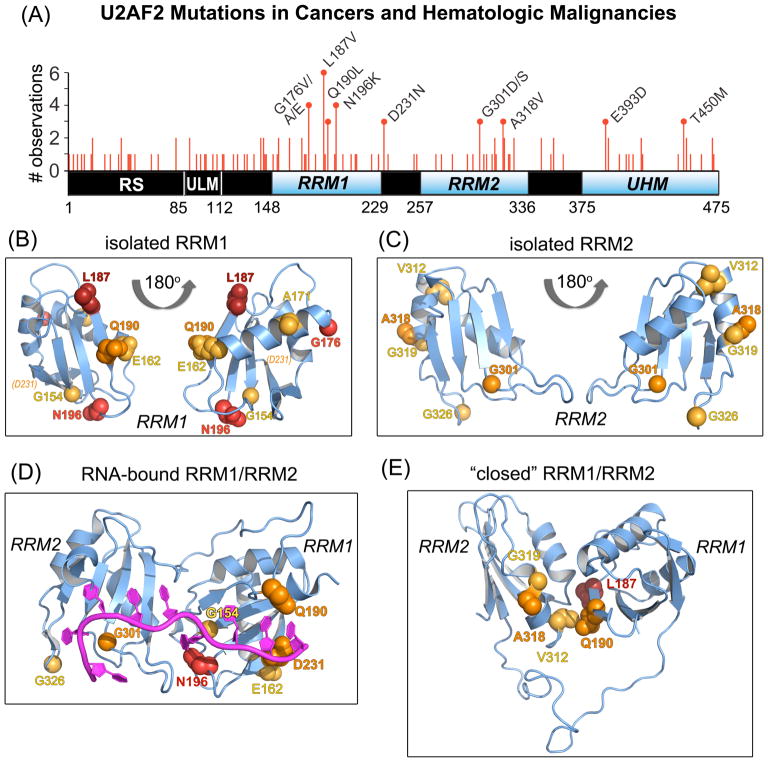
Figure 1.
U2AF2 missense mutations in neoplasms mapped on RRM structures. (A) Non-redundant U2AF2 missense mutations in cancer genomes mapped on the protein domains. Line heights are proportional to the number of independent observations for each mutation summed among independent patient samples. Amino acids that are changed in three or more samples are labeled. RNA recognition motifs (RRM1 and RRM2) are blue and other domains black. Residue numbers of domain boundaries are given below the schematic diagram. RS, arginine-serine-rich; ULM, U2AF ligand motif; UHM, U2AF homology motif. (B–C) Crystal structures of U2AF2 RRM1 (B) and RRM2 (C) at 1.07 and 1.11 Å resolutions. Views are rotated 180° about the y-axis. An affected residue (D231) located beyond the C-terminus of the crystallized construct (P229) is indicated in italicized parentheses. (D) The RNA-bound, “open” U2AF2 conformation (PDB ID 5EV4). (E) A typical “closed” U2AF2 conformation in the absence of RNA (PDB ID 2YH0). For clarity, a single representative of the NMR/PRE-fitted ensemble is shown in (E). Affected residues in (B–E) are space-filling atoms colored by the number of non-redundant mutations: 5, firebrick; 4, red; 3, orange; 2, yellow-orange.