Figure 3. TINAT-exon fusion transcripts encode novel protein isoforms with abnormal functions.
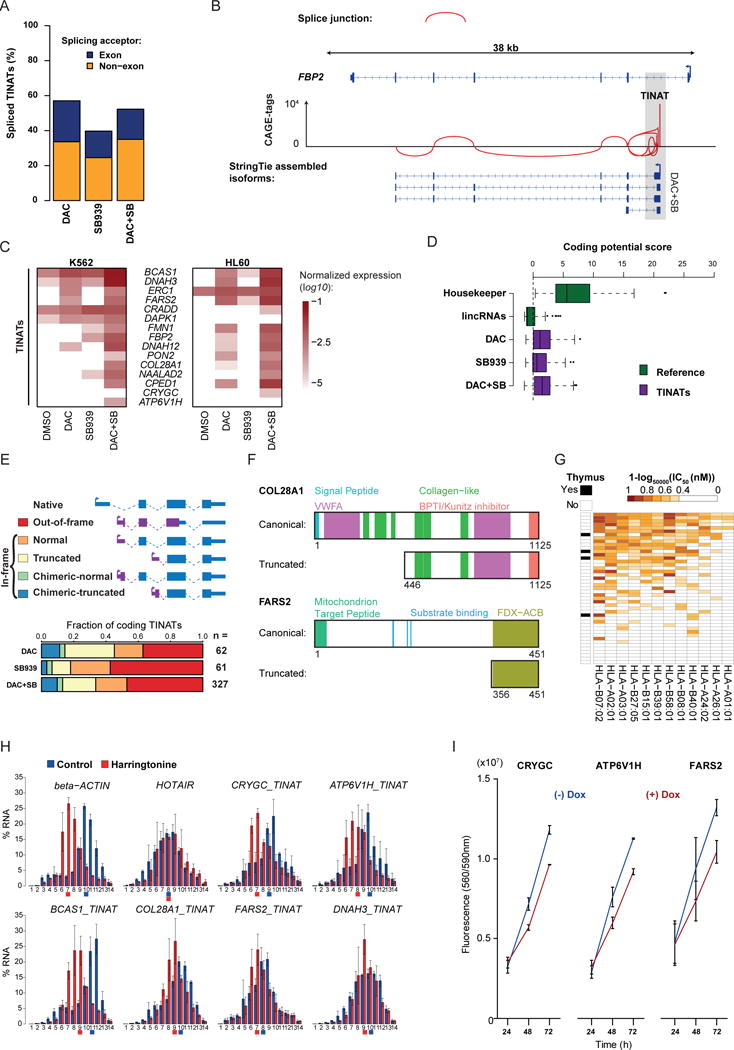
a) Fraction of TINATs having > 1% split CAGE-seq reads
b) Splice junctions at the FBP2 locus based on TINAT-derived CAGE-tags of DAC+SB treated NCI-H1299 cells.
c) TINAT-exon fusion transcript expression in K562 (left) and HL60 (right) cells. The log10 of the mean expression from three independent experiments relative to housekeepers is shown.
d) The coding potential of 100 housekeeping genes, 100 randomly selected ncRNAs, and TINATs was assessed using the coding potential calculator51. Dashed line denotes the threshold for protein-coding transcripts.
e) Schematic representation of the different scenarios for the translation of TINAT-exon fusion transcripts (upper panel). ORFs were categorized based on the criteria described in the online methods. The canonical (blue) and the novel, TINAT-derived sequence (purple) are schematically shown. Bottom panel depicts fraction of TINATs in each category.
f) COL28A1 and FARS2 protein domains for the canonical and truncated isoform are illustrated. Numbers below proteins indicate amino acid positions.
g) NetMHCpan52 was used to predict the binding affinity of 12 major HLA alleles (columns) for 45 DAC+SB chimeric peptide sequences (rows). The presence of a TINAT within the adult thymus is displayed.
h) Distribution of beta-actin, HOTAIR, and five TINAT-exon fusion transcripts along polysome fractions. Colored squares below horizontal axis line indicate the fraction where half of the mRNAs have accumulated.
i) Cell viability of NCI-H1299 reporter cells transduced with DOX-inducible TINAT-derived ORFs with or without DOX. Data from two independent experiments are shown.
