Figure 5. TINATs arise from long-terminal repeats especially of the LTR12 family.
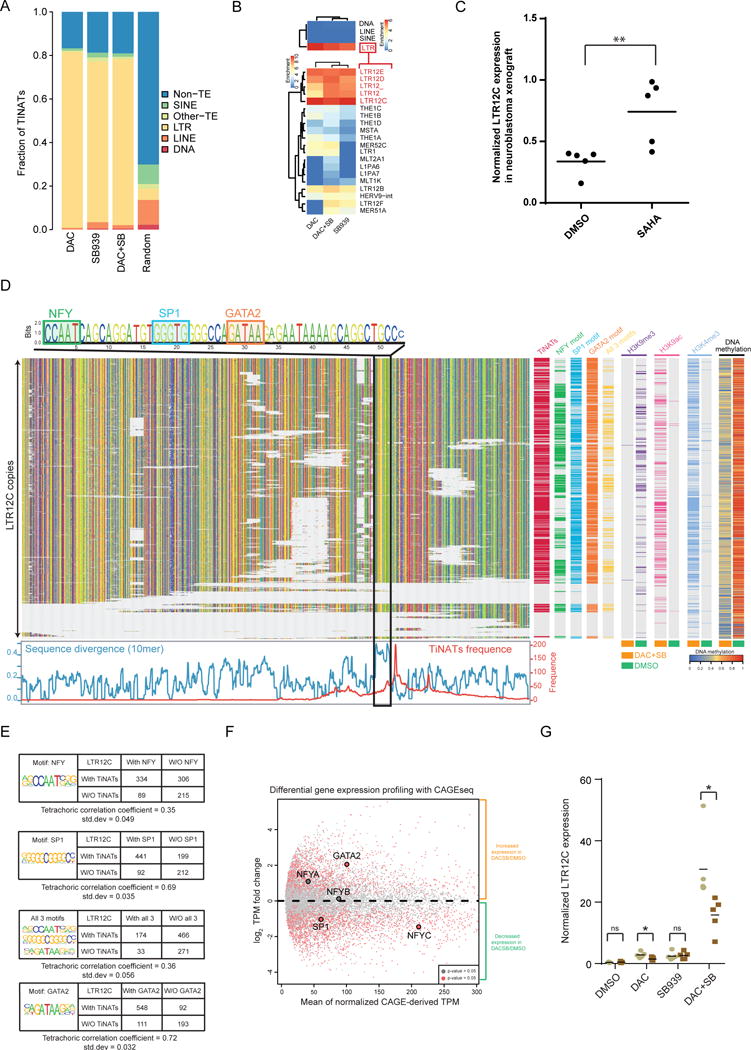
a) TINATs overlapping with transposable element (TE) classes.
b) Cluster analysis of enrichment scores for TINATs across TE classes (top panel) and LTR families (bottom).
c) qRT-PCR expression analysis of LTR12C copies relative to housekeepers in BE(2)-C neuroblastoma cells xenotransplanted into mice treated with DMSO or SAHA. Vertical lines represent the mean.
d) Sequence alignment of LTR12C copies. G, A, T, C, nucleotides are colored by yellow, green, red, and blue, respectively (top left). TF motifs are highlighted. TINAT frequency and sequence divergence between LTR12C copies with and without TINATs is shown below. Right panel displays the presence of TINATs, TF motifs, histone modifications, and DNA methylation.
e) Association between TINAT expression and the presence of NF-Y, SP1 and GATA2 motifs. Enrichment of SP1 and GATA2 sites in LTR12C with TINATs was significant (Pearson’s chi-squared test, P < 2.2e-16).
f) Differential gene expression between DAC+SB and DMSO using CAGE peaks. NF-Y, Sp1, and GATA2 are highlighted. NF-Y transcription factor is a trimeric complex of NYFA, NFYB and NFYC. Genes with significant expression differences are labeled in red (t-test p-value < 0.05).
g) Expression of LTR12C copies relative to housekeepers in the presence (brown) and absence (grey) of siRNAs targeting GATA2. Data from five independent experiments are shown. The mean from five independent experiments is shown.
