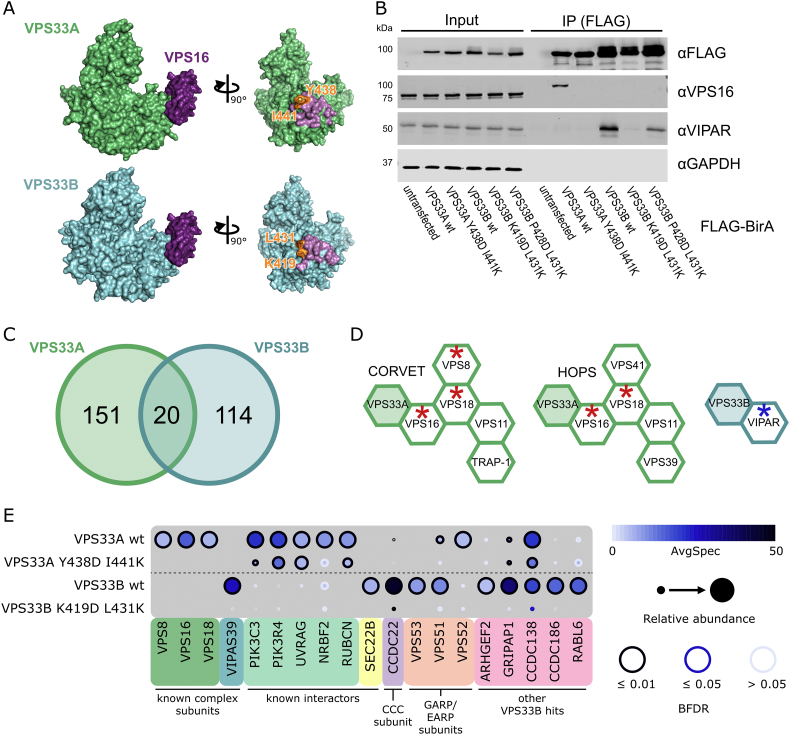
Fig. 1.
BioID results for VPS33A and VPS33B. (A) Crystal structure of VPS33A in complex with VPS16 residues 642–736 (top) and a homology model of VPS33B docked onto this complex (bottom). Residues shown in orange are mutated in panel B. Molecular graphics were generated using PyMOL (Schrodinger LLC). (B) HEK293 Flp-In T-REx cells were stably transfected with VPS33A and VPS33B (wild-type and mutant) constructs with C-terminal FLAG-BirA* tags and expression was induced with tetracycline. After immunoprecipitation (IP), samples were immunoblotted for the indicated endogenous protein. (C) Distribution of proteins identified by BioID with a BFDR of ≤ 0.01 and at least twofold enrichment in the wild-type sample over the negative-control mutant sample. (D) CORVET, HOPS and VPS33B–VIPAR (CHEVI) complexes, with subunits identified in BioID results marked with red asterisks (VPS33A as bait) or blue asterisk (VPS33B as bait). (E) Selected BioID results, shown as dot plots [59]. The spectral counts for each indicated prey protein are shown as AvgSpec. VIPAS39 = VIPAR, RUBCN = RUBICON. A complete list of the proximal proteins for each bait is available in the supplementary data file.