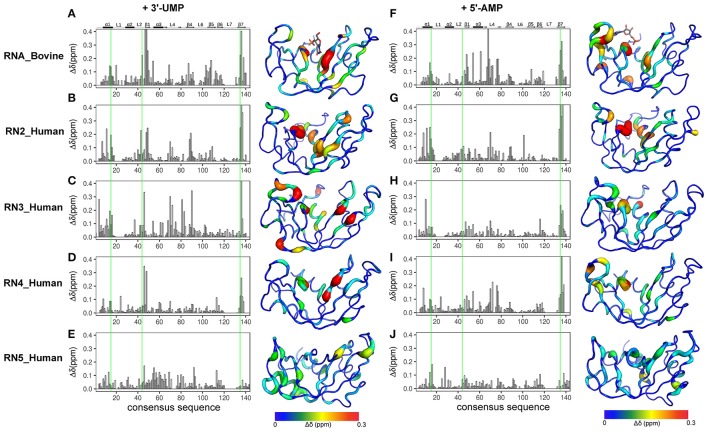
Figure 3.
Effect of 3′-UMP and 5′-AMP ligand binding on functionally distinct RNases. Compounded chemical shift perturbations (Δδobs at the highest ligand concentration) relative to the apo form upon binding of two mononucleotides 3′-UMP (A–E) and 5′-AMP (F–J) for bovine RNase A (A,F) and human RNases 2 (B,G), 3 (C–H), 4 (D,I), and 5 (E,J) are plotted as function of consensus sequence. Active-site residues (His12, Lys41, His119, RNase A numbering) are highlighted using green lines. Chemical shift perturbations were calculated by comparing the shifts at the largest enzyme:ligand molar ratios relative to the apo state for each enzyme. Δδobs are shown using the putty representation on the 3D structures to the right of the plots. Secondary structure of bovine RNase A is traced on top of the sequence alignment using blocks, arrows and dashed lines to represent α-helix, β-strand and loop regions, respectively. The 3′-UMP and 5′-AMP ligand positions, depicted on the structures of RNase A (A,F), were obtained from PDB entries 1O0N and 1Z6S, respectively.