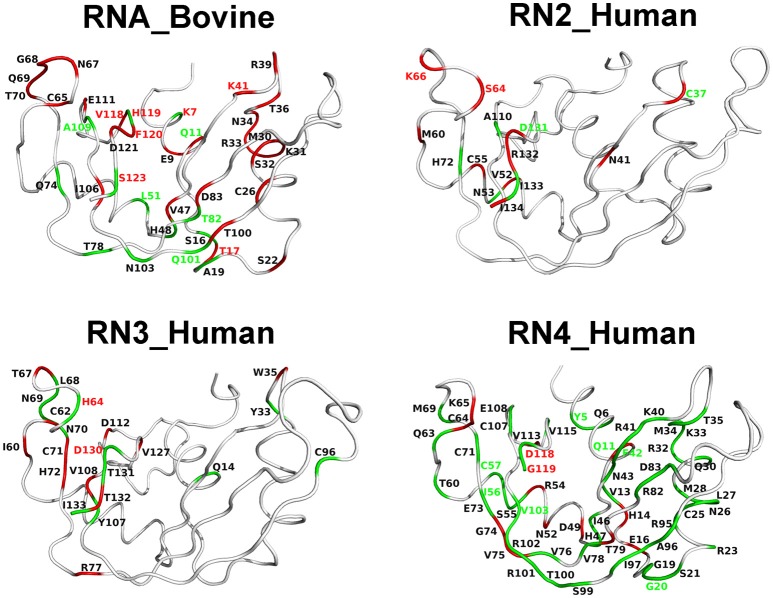
Figure 6.
Coordinated conformational exchange and chemical shift perturbations in functionally distinct RNases upon ligand binding. Millisecond timescale dynamics was probed using 15N-CPMG relaxation dispersion experiments following binding of 3′-UMP and 5′-AMP to bovine RNase A (RNA_Bovine) and human RNases 2 (RN2_Human), 3 (RN3_Human), and 4 (RN4_Human). RNase 5 (RN5_Human) is not shown, as no measurable relaxation dispersion effects were observed upon ligand binding to this enzyme on this timescale. Residues displaying changes in conformational exchange upon binding (relative to the apo state) for both 3′-UMP and 5′-AMP are shown in green, while residues that show dynamical changes upon binding of only one of the two ligands are shown in red (see section Methods). Dynamical residues that also display coordinated or uncoordinated displacements in the chemical shift projection analysis (Figure 5) are highlighted using green and red residue labels, respectively.