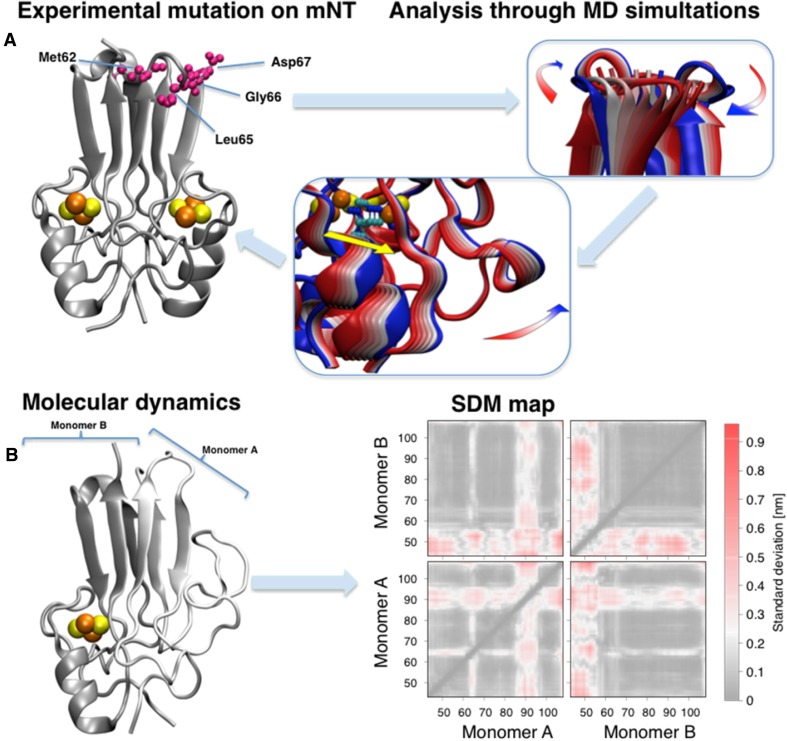
Fig. 6.
mNT modifications and theoretical analysis. (a, left) Crystal structure of mNT [25] highlighting the allosteric mutated residues at the top of the β-cap (violet). All crystal structures are available of the mutated proteins [51]. The β-cap mutations alter the coordinated motions of the domain (a, center panel), correlated with the flexibility of the cluster binding domain (right) and, in particular, with the coordinating histidine (yellow colored arrow) [51]. The colors of the cartoon structure in the central and right panels span from blue to red representing the movement along the principal vibrational mode of the protein. (b, left) Representative structure of the mNT in absence of one [2Fe-2S] cluster from monomer A obtained using replica exchange molecular dynamics [52]. (b, right) The effects of the cluster absence on standard deviation maps [52]. Here each pixel represents the standard deviation of the distance between each residue couple. The regions which are mostly affected by the cluster absence are the α-helix of the monomer losing the cluster and the L1 domain of the other [85]