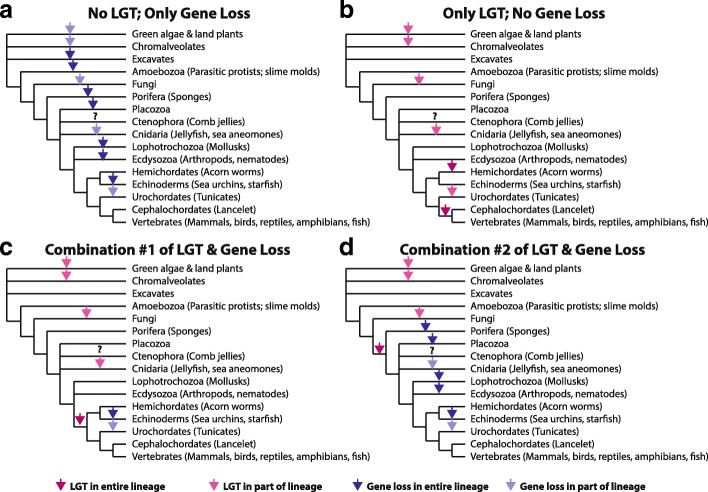
Fig. 3.
Schematics Illustrating Possible Paths to Explain the Current Distribution of ACY3/ASAP Homologues. Phylogenies from the Tree of Life Web Project were concatenated and LGT and gene loss events were overlaid in four possible scenarios: a presence of ASPA/ACY3 in the last common ancestor of eukaryotes and only gene loss, b absence of ASPA/ACY3 in the last common ancestor of eukaryotes and only LGT, c combination of LGT and gene loss where LGT occurred in the ancestor of all deuterostomes, d combination of LGT and gene loss where LGT occurred in the ancestor of all animals. Maroon arrows are used to indicate LGT in an entire lineage, while pink arrows are used to indicate LGT in a subset of taxa represented here. Dark purple arrows are used to indicate gene loss in an entire lineage, while lavender arrows are used to indicate gene loss in a subset of taxa represented here. It is not possible at this time to determine the likelihood of all of these possible scenarios, without better resolution of the eukaryotic tree of life, more sequence data from non-animal lineages, and a better understanding of the rates of gene loss and LGT in eukaryotes, which likely vary by lineage. However, it seems improbable that gene loss alone explains these results, which suggests that some LGT from bacteria to eukaryotes, and most likely, animals is responsible for the distribution of ACY3/ASPA homologues observed today