Figure 3. Crystal structure of an interdomain-locked HIV-1 Env trimer.
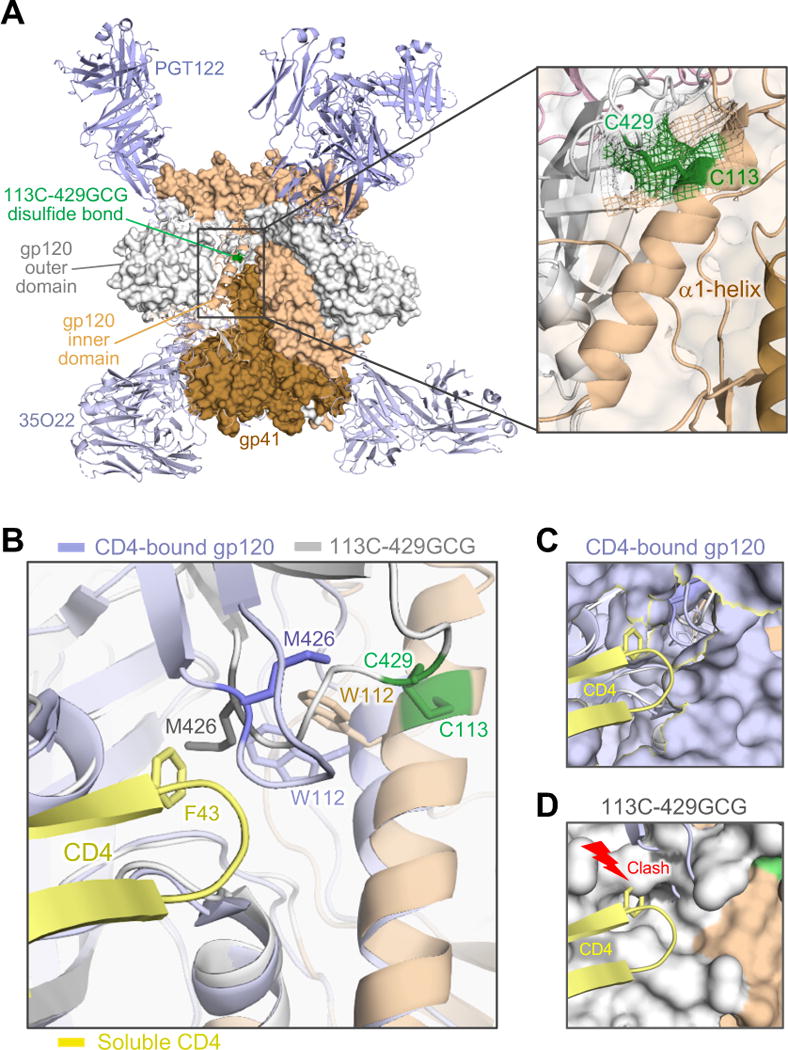
(A) The BG505 SOSIP.664 113C-429GCG trimer, bearing an engineered neo-disulfide bond between residues 113 and 429, was crystallized in complex with bNAbs PGT122 and 35O22. The overall structure, solved at 4.3 Å, is shown as protein surface with the α1-helix (beige) and the β20-21 loop (grey) of one protomer in cartoon representation. The neo-disulfide bond linking the two domains is highlighted in green. The inset shows a close-up of the stabilizing disulfide bond. Electron density (2Fo-Fc contoured at 1σ) clearly confirms the disulfide linkage.
(B) Overlap of the stabilized trimer structure (beige/grey) with a CD4-bound gp120 structure (light blue; 1RZJ). The F43 loop of CD4 (yellow) is shown aligned with the disulfide-stabilized trimer. Several residues in the hydrophobic pocket of CD4-bound gp120 are reoriented, including M426, W427 and W112. As in other CD4-bound structures (Huang et al., 2005; Kwong et al., 2000; Kwong et al., 1998; Pancera et al., 2010), including the recently reported structure of a CD4-bound SOSIP.664 trimer (PDB ID: 5VN3) (Ozorowski et al., 2017), Met426 is oriented away from the F43-binding pocket. In contrast, the disulfide bond precludes M426 from reorienting and constrains the distance of the β20-β21 loop from the α1-helix. (C) Surface representation of gp120 in CD4-bound conformation, with F43 finding its way into its hydrophobic binding pocket without interference from M426.
(D) In the locked trimer, M426 is positioned inside the pocket, as in other unbound trimer structures (4TVP, 5CEZ, 5FYL), creating a clash with the incoming F43. See also Figure S6.
