Fig. 3.
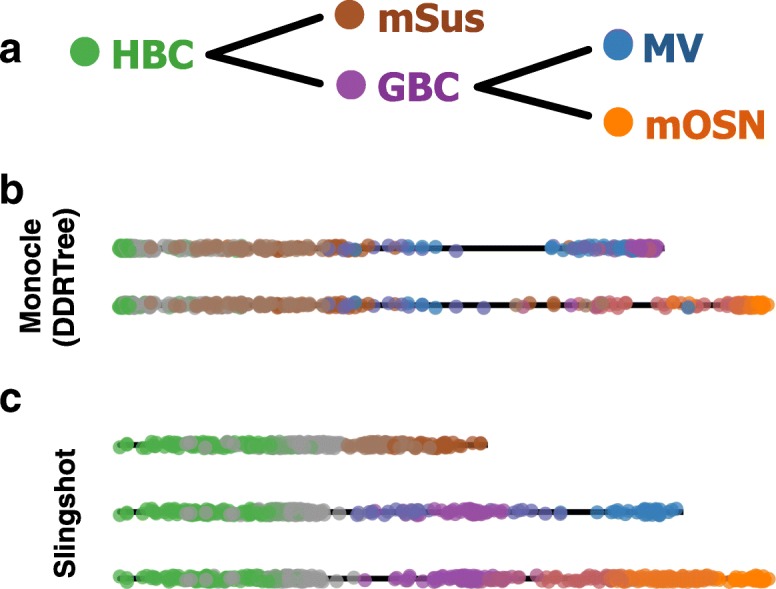
Multiple lineage inference: OE dataset. Pseudotime variables for each lineage inferred by Slingshot and Monocle 2 on the three-lineage OE dataset of [26]. Panel (a): Known biological relationships between cell types. Panel (b): For Monocle 2, we used the DDRTree algorithm to obtain a two-dimensional (or five-dimensional, see Additional file 1: Figure S3d) representation of the data and selected the starting state based on the highest percentage of cells from the HBC cluster. Panel (c): For Slingshot, we used the top five PCs and clustered cells by RSEC, as in the original article. The HBC cluster was specified as the origin and the mSus cluster as an endpoint; other endpoints were identified without supervision
