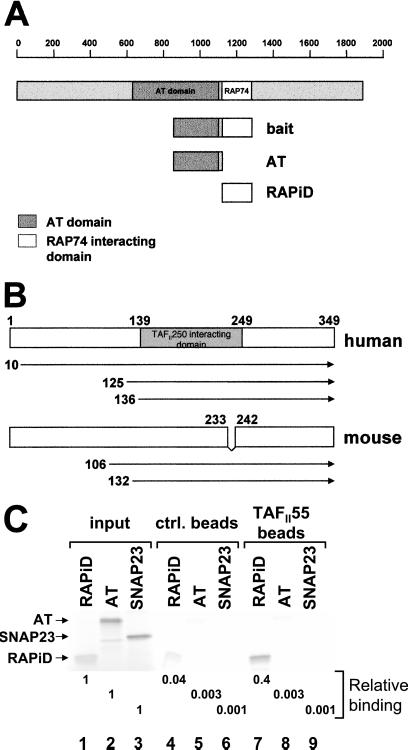
Figure 1.
TAFII55 binds TAFII250. (A) The TAFII250 fragment (amino acids 848-1279), spanning the AT and RAPiDs, was used as bait in a yeast two-hybrid screen to isolate TAFII55 clones. The location of the bait fragment, relative to the full-length molecule, is shown. TAFII250 AT domain (shaded box) is located approximately between amino acids 640 and 1093, which corresponds to the Drosophila AT domain (amino acids 612-1140) (18). (B) TAFII55 clones isolated in yeast two-hybrid screens. The locations of the five isolated TAFII55 clones are shown relative to the full-length human and mouse proteins. Although the clones differed in their 5′ termini, they shared a common 3′ end that extended to encode the carboxyl terminus of the TAFII55 protein. The mouse TAFII55 differs from the human in a deletion spanning amino acids 233–242. (C) TAFII55 binds to the RAPiD of TAFII250. Flag-tagged TAFII55 was used to assess its ability to bind to either [35S]methionine labeled, in vitro translated AT, or RAPiD domains, as shown in A (AT, lanes 2, 5, 8; RAPiD, lanes 1, 4, 7). In vitro translated SNAP23 protein was used as a control for nonspecific binding (lanes 3, 6, 9). The relative binding of the TAFII250 fragments to TAFII55 was quantitated and calculated relative to input, as shown beneath the lanes. Arrowheads mark the positions of input proteins.