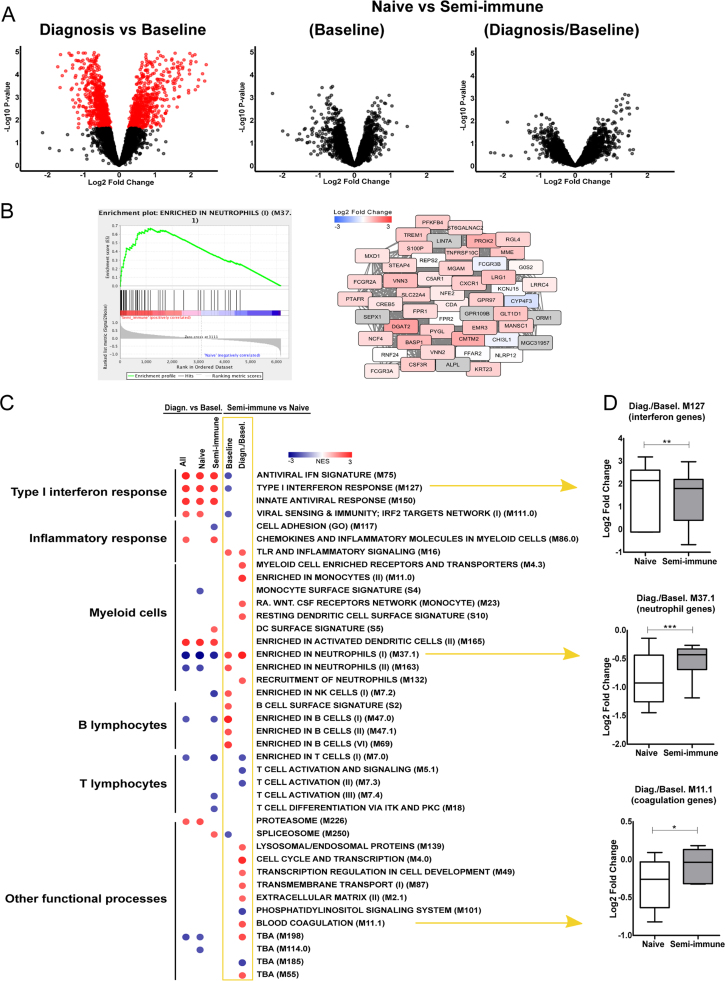
Fig. 3.
Blood Transcription Modules (BTM) analysis ofP. vivaxCHMI trial. Blood transcriptomes of six naïve and six semi-immune subjects were described previously. A, Gene-level analyses for all subjects between diagnosis and baseline levels; between naïve and semi-immune subjects at baseline, and at the time of P. vivax malaria diagnosis. Genes selected at FDR < 0.1 are colored in red. B, Representative gene set ranking plot and, network plot for BTM annotated as Enriched in Neutrophils (II) (M37.1). It shows the comparison between semi-immune and naïve subjects at the time of diagnosis. Blue to red scale in the network corresponds to gene expression fold changes of semi-immune/naïve subjects. Genes that were not measured in this experiment are colored in gray. C, Summary of BTMs associated with transcriptional profiles according to each statistical comparison. Comparison between semi-immune and naïve subjects at the time of diagnosis was based on baseline normalized intensities. Gene set enrichment analysis was used to identify significant associations at FDR < 0.05. The blue to red scale indicates negative or positive associations based on normalized enrichment scores (NES). D, Boxplots of highlighted BTMs comprising interferon genes (M127), neutrophil genes (M37.10, and blood coagulation genes (M11.1). Significance levels are shown as *, p < 0.05; **, p < 0.01; ***, p < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).