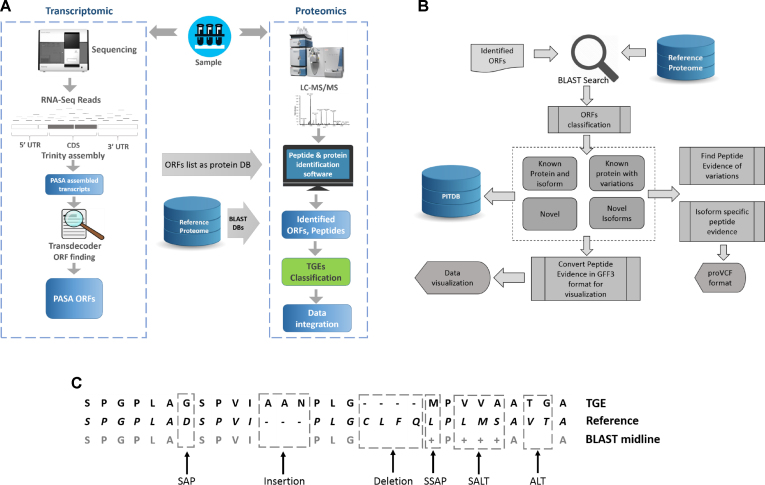
Figure 1.
PIT pipeline, now including classification of TGEs. (A) RNA-Seq assembly begins with the Trinity de novo transcript assembler. PASA is then used to assemble spliced alignments and identify alternative splicing events at transcript level. Transdecoder is used to predict ORFs from PASA’s transcripts. (B) TGEs are classified, based on sequence similarity to an existing proteome, into four main classes, (i) known protein or isoform, (ii) known protein or isoform with polymorphisms, (iii) novel isoforms and (iv) novel TGE. Within these main classes there are four polymorphism categories and sixteen novel isoform classes. We look for supporting peptides from the mass spectrometry (MS) data to verify these events at protein level. Variation information and peptide evidence for all the identified TGEs is output and deposited in a database called PITDB. (C) Nomenclature of polymorphism types derived from BLAST alignments used in this paper—see main text for details.