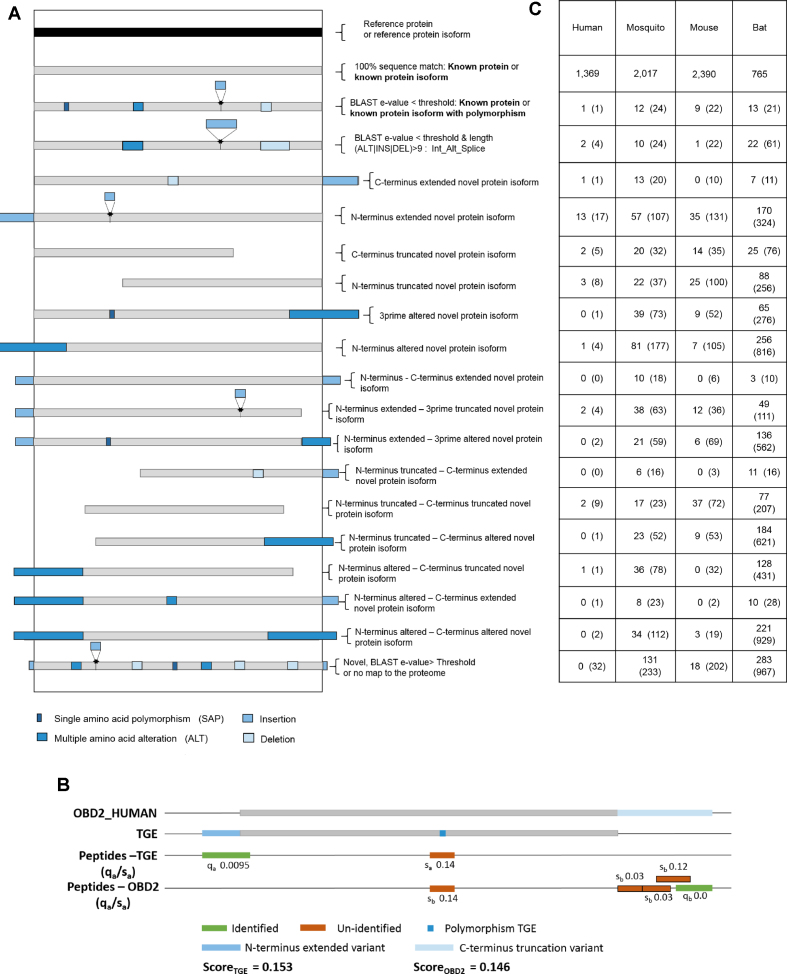
Figure 2.
(A) Classification of TGEs based on BLAST alignment to UniProt proteome (including isoforms) of the species under study. TGEs with 100% sequence map to UniProt proteins are labelled as known proteins or known isoforms. TGEs with BLAST e-value above 1 × 10−30 or that do not map to a UniProt protein are classified as novel. The remaining TGEs are classified into one of 17 types based on location, length and type of variation. (B) Example of scoring a putative novel isoform based on its mapping to the most homologous protein found by BLAST (ODB2_HUMAN). Only peptides not shared between the TGE and reference are used to compute the scores of the TGE and the reference protein, using Equation (1). Q-value scores and sample-specific detectability scores (SS) are used for identified and unidentified peptides respectively. In this case, the TGE score exceeds that of the reference sequence, suggesting we have a novel variant of ODB2. (C) Identified TGEs of each type supported by peptide evidence, and unique peptide evidence in parentheses. The proportion of novel findings is higher in species with less well annotated genomes.