Figure 1.
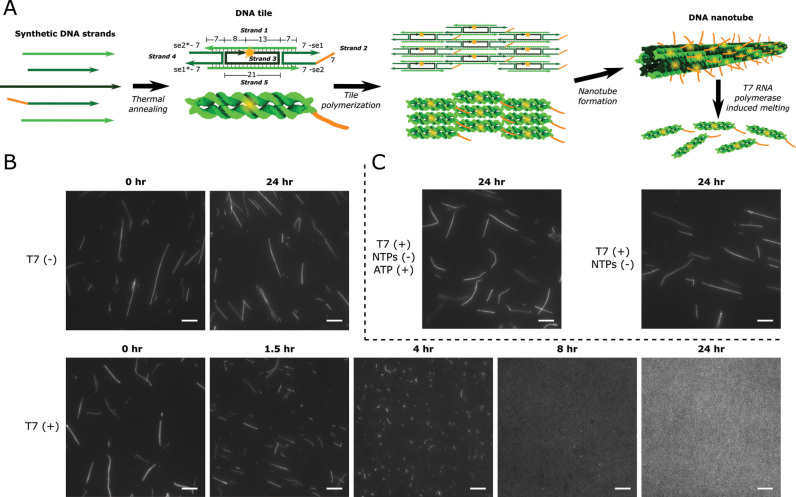
DNA nanotube design and disassembly in the presence of 8.6 U/μl T7 RNAP in transcription conditions. (A) DNA nanotube structure and assembly. Left: DNA tiles are DAE-E double crossover molecules (71) that assemble from 5 strands of synthetic DNA. Tiles consist of two DNA helices that are rigidly bound by two crossover motifs with four single-stranded sticky end (se) regions on the two helix ends. Sticky ends are complementary across the diagonal of the tile (complementary sequences denoted by *) (1). Tiles also contain a single-stranded overhang domain on strand 2 (orange) and a Cy3 fluorophore (yellow) at the 5′ end of strand 3. Numbers next to domains indicate domain lengths in number of bases. Middle: The complementary sticky ends specifically program the DNA tiles to form the lattice shown as an abstract line representation (top) and a 3D rendering (bottom). Right: The lattice cyclizes to form nanotubes (1). (B) Fluorescence micrographs of DNA nanotubes (var1_7—SI Section 1) before and after incubation with (T7 (+)) or without (T7 (–)) T7 RNAP in transcription conditions. (C) Nanotube stability (var1_7) in the presence of T7 RNAP with ATP (ATP (+)) as the only NTP or without any NTPs (NTPs (–)). Scale bars: 10 μm.