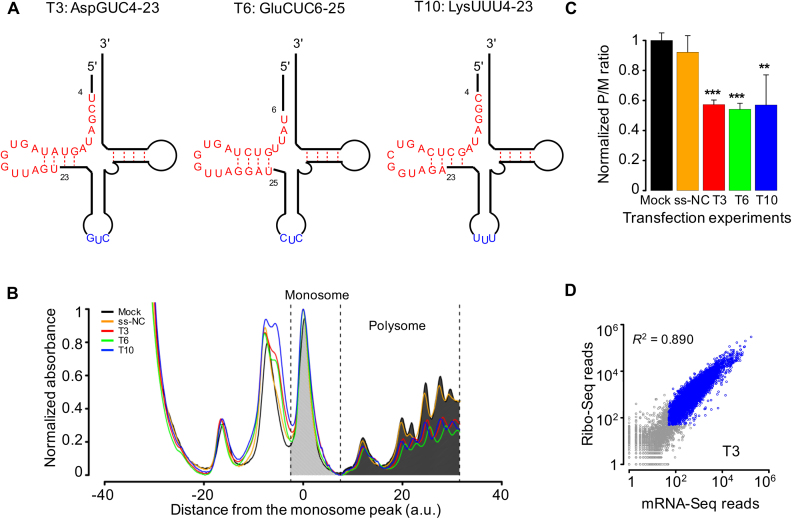
Figure 2.
Transfections of tsRNA mimics into S2 cells repress global translational activities. (A) Schematic representations of the structures of three tsRNAs tsRNAAspGUC (T3), tsRNAGluCUC (T6) and tsRNALysUUU (T10) in the mature tRNAs. tsRNAs are in red and the anti-codons in blue. (B) Absorbance profiles (at a UV wavelength of 254 nm) of RNAs partitioned using 10–45% sucrose gradients from S2 cells transfected without any RNA sequence (Mock, black), with a negative-control small RNA (ss-NC, orange), with tsRNA T3 (red), T6 (green) or T10 (blue). For each profile, both the absorbance and the position were normalized according to the monosome peak. Dashed lines indicate the boundaries of the monosome and polysome partitions for a measurement of the overall translational activity. (C) The ratios of the aggregate intensities within polysome partition (P) to the monosome partition (M) normalized by the median value of the control experiments. Error bars represent one standard deviation computed from three repeated experiments. Asterisks indicate significant different P/M ratios between transfection and mock: *P < 0.05; **P < 0.01; ***P < 0.001. (D) Scatter plots of the Ribo-seq read counts against the mRNA-seq read counts of individual genes in T3 transfection experiments. Blue circles represent genes that have >50 mRNA-seq reads and >50 Ribo-seq reads.