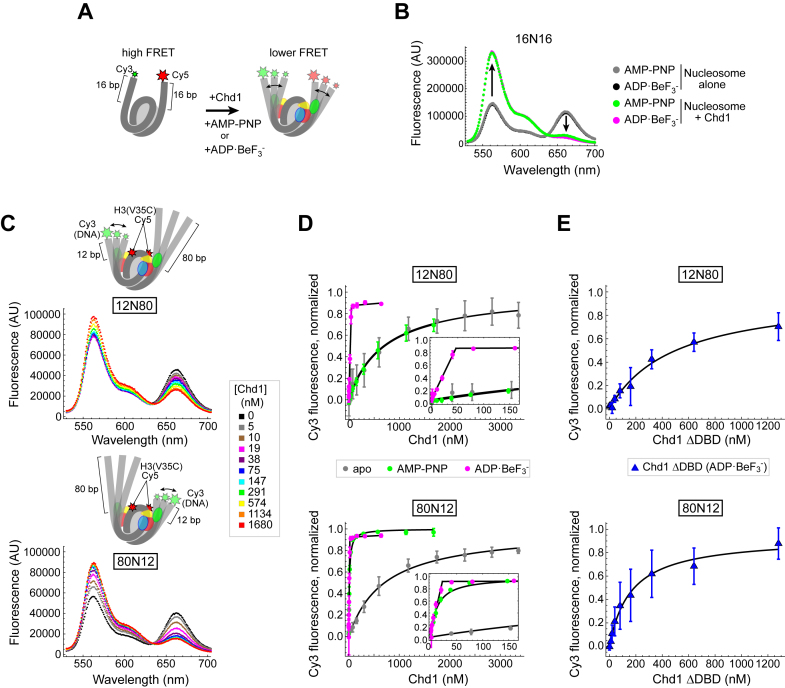
Figure 2.
Using FRET to monitor nucleosome unwrapping by Chd1. (A) Schematic illustration of Cy3/Cy5 FRET pair positioning on the 16N16 nucleosome, with one dye on the 5′ end of each DNA strand. Chd1 is colored by domains: chromodomains (yellow), ATPase motor (red/blue), and DNA-binding domain (green). (B) Wavelength emission scans for 10 nM 16N16 nucleosomes upon excitation at 510 nm (black, gray). As indicated by arrows, addition of 30 nM Chd1 promoted an increase in Cy3 emissions (564 nm) and a corresponding decrease in Cy5 emissions (664 nm) in the presence of AMP–PNP (green) and ADP·BeF3– (magenta). (C) Nucleosomes made with the Widom 601 positioning sequence are unwrapped by Chd1 more readily on the TA poor side. Cartoon representations of 12N80 (top) and 80N12 (bottom) are shown above wavelength emission scans (with 510 nm excitation) with increasing amounts of Chd1 in the presence of 1 mM AMP–PNP. (D) Nucleosome unwrapping monitored by Cy3 emission with excitation at 510 nm. Chd1 titrations were performed in the presence of 20 nM 12N80 (top) or 20 nM 80N12 (bottom) nucleosomes in various conditions: apo (gray), 1 mM AMP–PNP (green), and 1 mM ADP·BeF3– (magenta). Insets highlight the stoichiometric unwrapping response in ADP·BeF3– at low Chd1 concentrations. Error bars represent standard deviation from three or more replicates. Each dataset was fit to a binding isotherm, with the calculated averages shown by black lines. (E) The Chd1 DNA-binding domain (DBD) is not required for nucleosome unwrapping. Titrations of Chd1ΔDBD performed in the presence of 12N80 (top) and 80N12 (bottom) nucleosomes under ADP·BeF3– conditions.