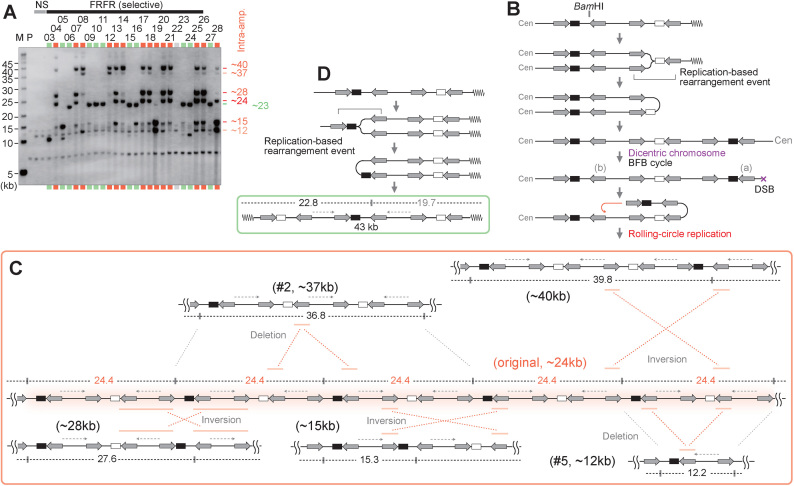
Figure 3.
Characterization of amplified chromosomal structures. (A) Southern blot of BamHI-digested DNA with the leu2d probe. The samples in Figure 1B–D, are digested with BamHI. The samples marked in red and green indicate intra- and extra-chromosomal products, respectively. The sample marked in light gray showed no sign of amplification, suggesting Leu+ recombination between the leu2d marker and the mutated original leu2 allele on chr III. M: S. cerevisiae marker; P: the parental strain, LS20; NS: non-selective conditions. (B) A predicted process for intra-chromosomal amplification. The replication-based event forms a dicentric chromosome followed by BFB cycles, producing a chromosomal break that can initiate RCR. (C) Predicted structure of RCR-amplification products associated with intensive rearrangements. RCR-amplification could form an original repeat array that produces 24.4-kb BamHI-fragments. RCR-associated inversions and deletions produce rearranged BamHI-fragments (12.2, 15.3, 27.6, 36.8, 39.8 kb). IRs engaged in the inversions are marked with red bars. (D) A predicted process for extra-chromosomal amplification. The replication-based rearrangement event between IRs marked with a bracket forms a 43-kb extra-chromosome. The BamHI-map indicates a leu2d-containing fragment (black) and a non-hybridizing fragment (gray).