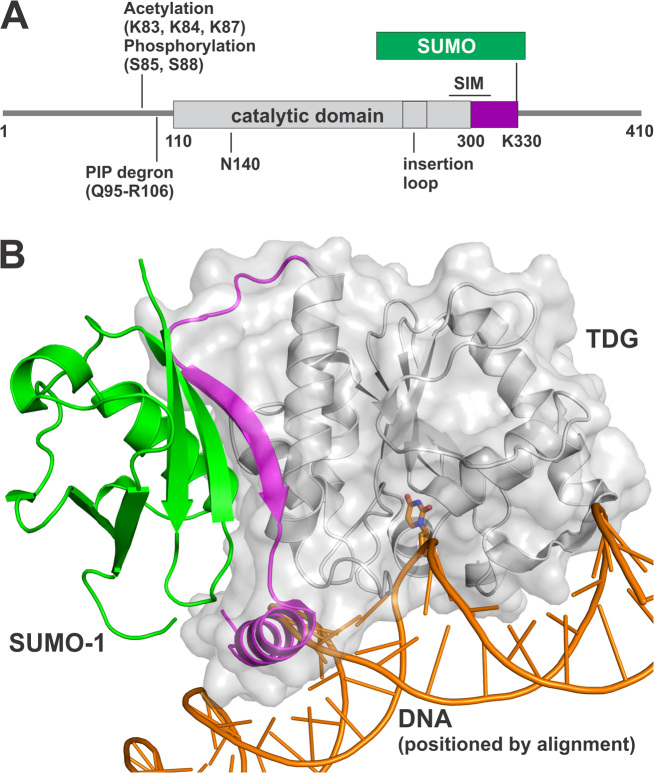
Figure 1.
SUMO modification of TDG. (A) Primary structure of TDG including the catalytic domain and two flanking disordered regions that mediate various functions. Shown are post-translational modification (PTM) sites, the SUMO-interacting motif (SIM), and the PIP degron motif that enables TDG interaction with PCNA, ubiquitin modification, and degradation. The site(s) of ubiquitin modification is currently unknown. (B) Crystal structure (2.1 Å) of the TDG catalytic domain (residues 112–339) modified by SUMO-1 (green) (PDB ID: 1WYW); residues 301–331 of TDG (magenta) exhibit some secondary structure (β-strand, α-helix) for sumoylated TDG but are likely disordered in unmodified TDG. Because no structure is available for sumoylated TDG bound to DNA, the DNA shown here (orange) was positioned by aligning a structure (1.54 Å) of DNA-bound TDG (PDB ID: 5HF7, hidden) with that of TDG∼SUMO-1 (shown), using PyMol (RMSD = 0.573 Å for 136 Cα atoms). While this model cannot be accurate, because the SUMO-induced helix of TDG overlaps with the DNA, it is useful for illustrating the location of the SUMO–SIM interface relative to the DNA binding region and the active site.