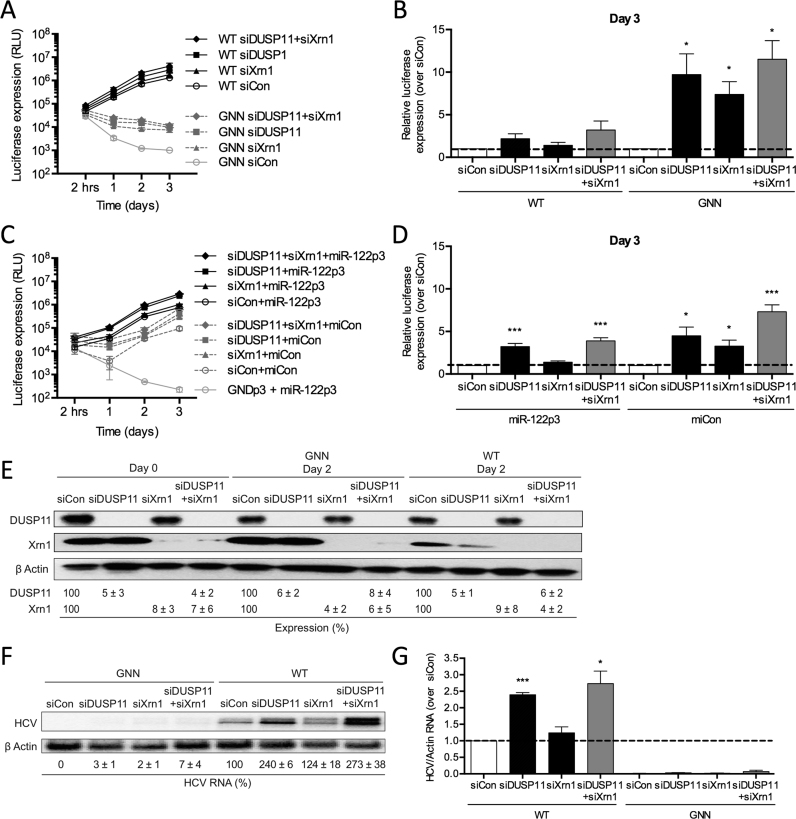
Figure 9.
miR-122 protects the HCV genome from the pyrophosphatase DUSP11 and subsequent 5′ decay mediated by Xrn1. (A) Huh-7.5 cells were electroporated with siDUSP11, siXrn1 or siControl (siCon) at day –2 and at day 0 cells were electroporated again with the indicated siRNA, FL WT or GNN HCV RNA, and a firefly luciferase control mRNA. Replication was measured by evaluating luciferase production at the indicated timepoints and is quantified at day 3 for FL HCV compared with siCon (B). (C) Huh-7.5 cells were electroporated with siDUSP11, siXrn1 or siCon at day –2 and at day 0 cells were electroporated again with the indicated siRNA, S1+S2p3 SGR or S1+S2p3 GND SGR, a Renilla luciferase control mRNA, and miR-122p3 (miR-122-dependent) or miCon (miR-122-independent). Replication was measured by evaluating luciferase production at the indicated timepoints and is quantified at day 3 for S1+S2:p3 SGR compared with siCon in (D). (E) Western blot showing knockdown efficiency with antibodies against DUSP11, Xrn1 and β-actin. Percent knockdown ± standard deviation relative to siCon in indicated. F) Northern blot analysis of FL HCV RNA accumulation during DUSP11 and Xrn1 knockdown at day 3. (G) Densitometry quantification of Northern blot analysis of FL HCV RNA accumulation normalized to siCon. All data are representative of at least three independent experiments and statistical significance was determined by paired parametric t test.