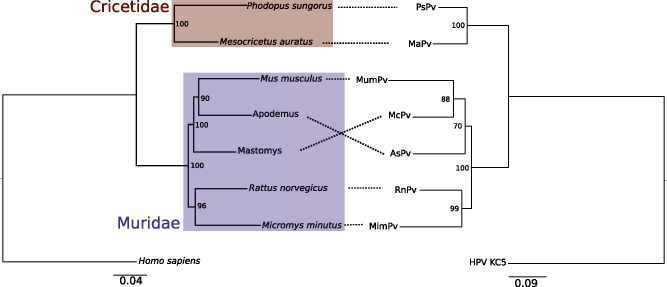
Figure 2.
Phylogenetic congruence of papillomaviruses (right) and their hosts (left). Relationships between host and virus are indicated by spotted black lines. The host phylogeny was inferred with same loci and methods as in Figure 1, while the PV phylogeny is based on conserved regions of E1, L1, L2 with the third codon position stripped. Both trees were inferred with RAxML 8.2.9. Number at nodes indicates bootstrap support based on one hundred replicates. Branch lengths are in expected number of nucleotide substitutions per site. For the PV phylogeny a related human PV (GenBank accession NC_026946) was picked as the outgroup. On the host phylogeny, colored boxes indicate families of rodents from the superfamily Muroidea.