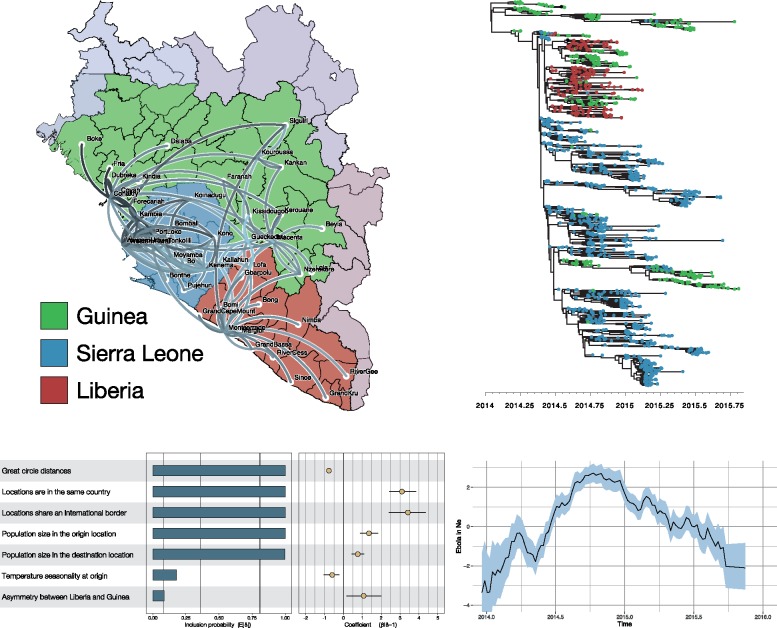
Figure 1.
Phylodynamic analysis of the 2013–2016 West African Ebola virus epidemic, encompassing simultaneous estimation of sequence and discrete (geographic) trait data with a GLM fitted to the discrete trait model in order to establish potential predictors of viral transition between locations. Plotted are a snapshot of geographic spread using SpreaD3 (Bielejec et al. 2016b), the maximum clade credibility tree, the posterior estimates of the GLM coefficients for seven possible predictors for Ebola virus spread (Bayes Factor support values of 3, 20, and 150 are indicated by vertical lines) and the effective population size through time, estimated by incorporating case counts.