Figure 1.
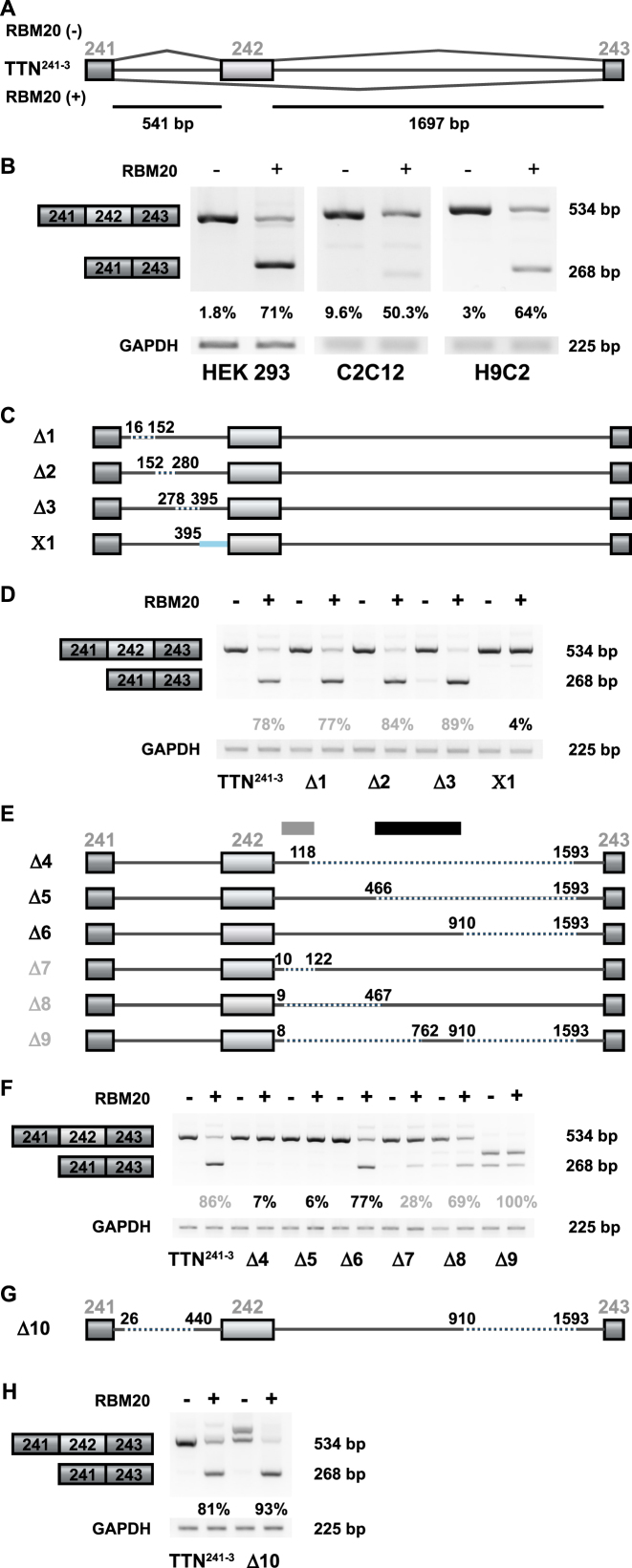
Human titin-derived splicing reporters respond to RBM20-mediated splicing repression in cell culture. (A) The splicing reporter TTN241-3 contains the human titin (TTN) genomic sequence from exon 241 to 243. Exon 242 is excluded in the presence of RBM20. Alternative exons are represented as light boxes, constitutive exons as dark boxes and introns as lines (bp = base pairs). (B) Validation of RBM20-dependent alternative splicing by RT-PCR. Transient transfection of HEK293, C2C12 and H9C2 cells with the TTN241-3 and empty vector (−) or a plasmid expressing human RBM20 (+) produces the expected RT-PCR products—with and without the alternative exon 242 (top and bottom band, respectively). Quantification of three independent transfections are shown as percentages of mRNA excluding the alternative exon. (C) TTN241-3 deletion mutants, generated to identify the RBM20-responsive splicing repression element in the upstream intron. Dotted lines represent deleted sequences and the blue line a sequence replaced with the first intron of the human β-globin gene. (D) Analysis of RBM20-dependent alternative splicing by RT-PCR in HEK293 cells transiently transfected with each reporter construct, as indicated. The RT-PCR product at 268 bp represents mRNA species that does not contain exon 242. Ratio of exon inclusion/exclusion was reduced to 4% only in the X1 replacement construct, while deletion in construct Δ3 enhances the RBM20 effect (quantification of three independent transfections). (E) TTN241-3 deletion mutants generated to identify the RBM20-responsive splicing repression element in the downstream intron. Dotted lines indicate deleted sequences. (F) Analysis of RBM20-dependent alternative splicing of deletion constructs affecting the downstream intron of TTN241-3. Constructs where deletions extend into the mid intronic region (Δ4 and Δ5, indicated with a gray box) reduce their repressor activity to <10% and Δ7 to <30%. Presence of the mid intronic region (indicated with a black box, construct Δ6) restores repressor activity (quantification of three independent transfections). (G) TTN241-3 deletion mutants generated to integrate the information on splicing responsive elements and truncate the minigene for subsequent analysis. Thin (gray) lines indicate TTN sequences, dotted lines indicate deleted sequences. (H) Analysis of RBM20-dependent alternative splicing of deletion construct affecting both the upstream and the downstream introns of TTN241-3. RBM20 responsiveness is maintained in construct Δ10 that lacks >50% of the downstream intron sequence (quantification of three independent transfections).