Figure 6.
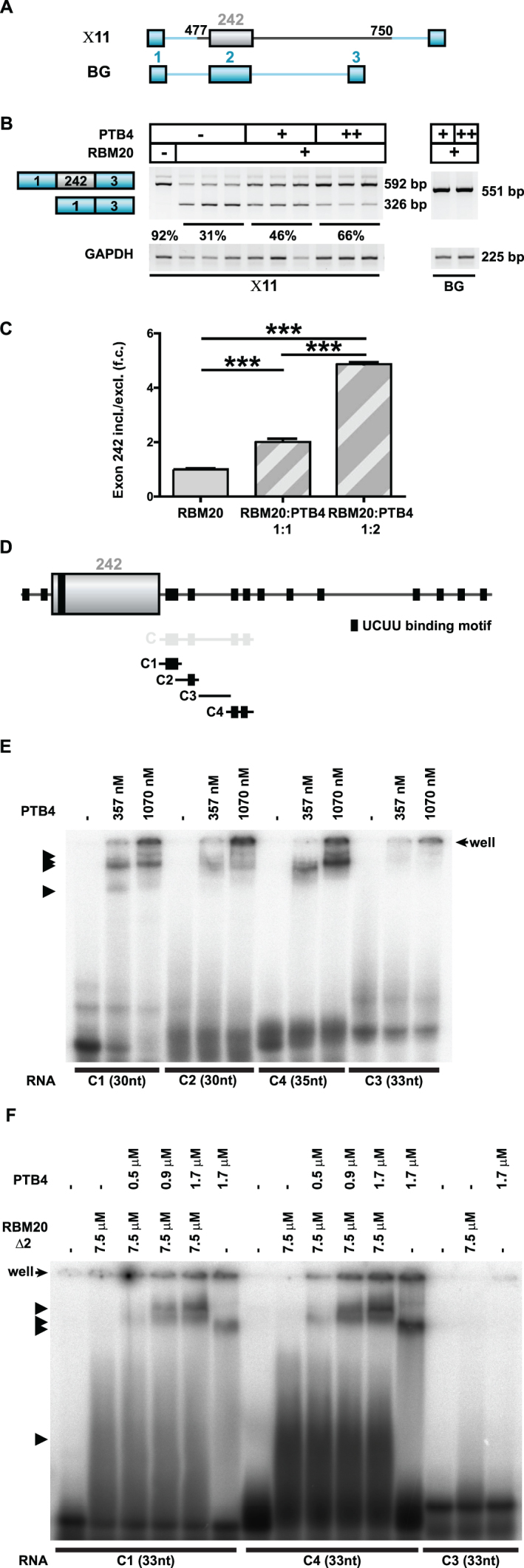
PTB4 enhancers TTN exon 242 inclusion dependent on the downstream intron. (A) Schematic representation of the human TTN (gray)/β-globin (blue) hybrid splicing reporters used to evaluate PTB4 effect on titin splicing regulation. (B) PTB4-mediated changes in the alternative splicing of the human titin/β-globin hybrid splicing reporter X11Changes in X11 alternative splicing were assayed in the background of RBM20. Increasing concentrations of PTB4 reverted the repressor activity of RBM20 on X11 splicing reporter. Titin-derived sequences missing splicing reporter (BG) was not responding to RBM20 or PTB4 (indicated on the right). (C) qRT-PCR (SYBR Green) quantification of PTB4-regulated alternative splicing events in the titin/β-globin mutant splicing reporter: n = 3; ***P < 0.001. (D) Schematic representation of the RNA transcripts used to study PTB4 binding. Black boxes indicate UCUU binding motifs. C1 transcript contains two UCUU binding motifs, organized in tandem, C2 contains one UCUU binding motif, C3 contains no UCUU binding motif and the C4 transcript contains two separated UCUU binding motifs. (E) RNA–protein complex formation as analyzed in a gel-shift assay. Black arrows indicate the observed formation of RNA–protein complexes. PTB4 binds all transcripts except for C3 that does not contain the consensus site. (F) RNA–protein complex formation analyzed by gel-shift assay using RBM20Δ2 and PTB4 protein. Black arrows indicate the formation of RNA–protein complexes. Increasing the concentration of PTB4 promotes the generation of additional RNA/protein complexes and leads to a reduction of RBM20Δ2/RNA complexes.
