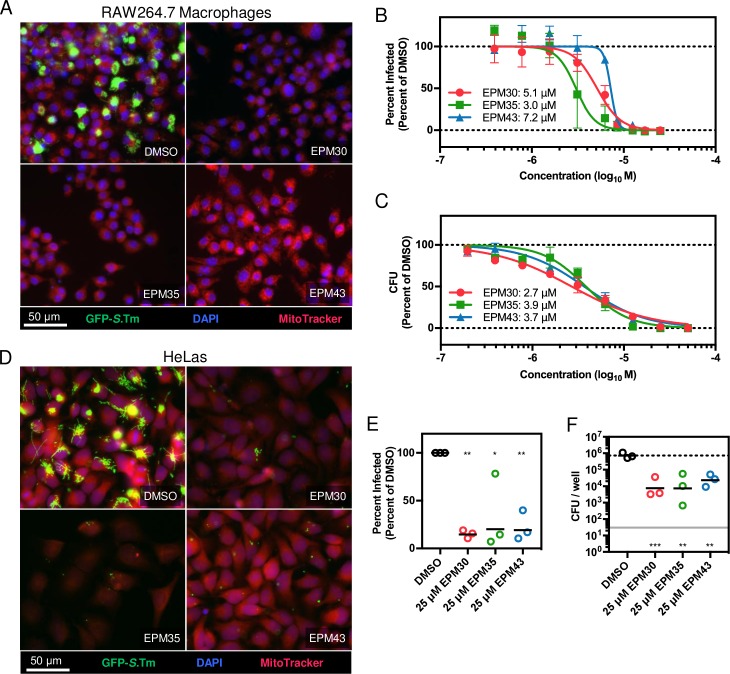
Fig 3. The three hit compounds decrease bacterial load of Salmonella in mammalian cells.
(A-C) Monitoring of bacterial load by GFP (SAFIRE) or CFU in RAW264.7 cells. (A) Representative micrographs of cells in 96-well plates infected with sifB::GFP Salmonella. Two hours after infection cells were treated with the indicated compound [25 μM] for 16 hours. (B) Dose response curve for SAFIRE and (C) CFU; keys includes IC50 values. (D-E) Monitoring of bacterial load by GFP (SAFIRE) in HeLa cells infected with Salmonella expressing rpsM::GFP and treated with the indicated compound [25 μM]. (F) Monitoring of bacterial load by CFU in BMDMs treated with compounds [25 μM]. Mean and SEM from three independent biological replicates. The nonlinear curve fitting (B, C) is constrained using uninfected cells as the minimum and DMSO-treated cells as the maximum. (E, F) * p < 0.05; ** p < 0.01, *** p < 0.001 compared to DMSO by one-way ANOVA with Dunnett’s post-test.