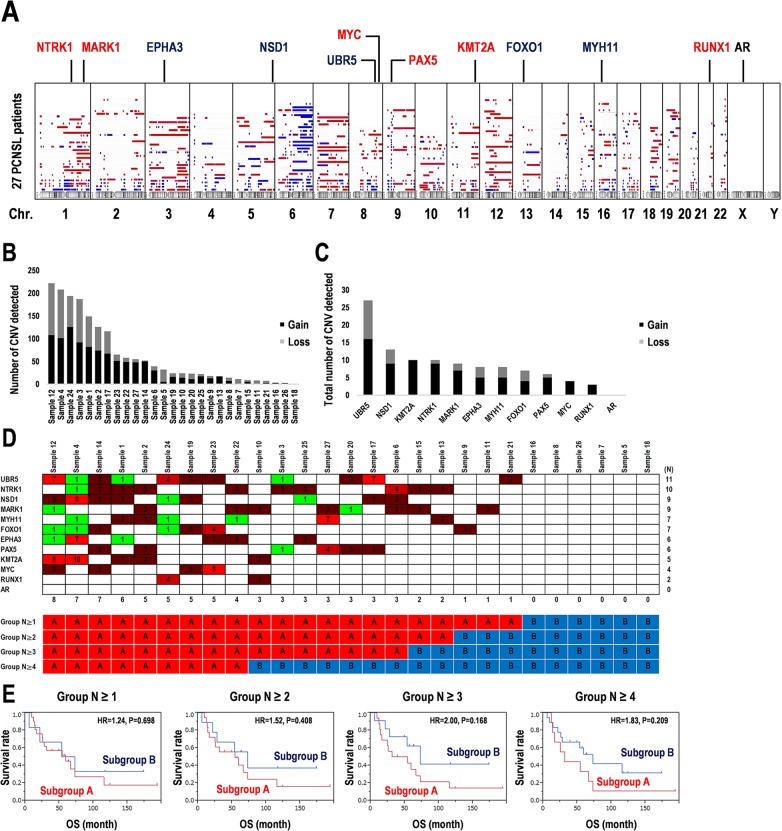
Figure 5. Copy number variations detected in 12 prognostic marker candidates in 27 PCNSL specimens.
(A) Copy number variations (CNVs) over the entire genome were visualized in karyotype view. Genomic positions of 12 candidates of prognosis markers were mapped. The regions of CNVs with gain (red) and loss (blue) indicate as colored bars and gene names. (B) Total numbers of the regions detected with significant CNVs in each sample. (C) Total numbers of the regions detected with significant CNV regions including the 12 candidate of prognosis markers. (D) CNVs detected into the region including the 12 candidate prognostic markers in each PCNSL specimen. Numbers in the matrix indicate CNV > 2: Gain (red) and < 2: Loss (green) (upper). Numbers at the bottom of the upper panel indicate the numbers of genes associated with CNVs in each PCNSL specimen. The 27 PCNSL specimens were divided into the two subgroups according to the CNV frequency per specimen after two-way clustering (lower). (E) Kaplan-Meier survival analysis for the combination of the 12 candidate genes associated with SNVs and CNVs. Analyses were performed according to the groups as shown in D. OS; overall survival time, HR; hazard ratio.