Figure 4. Overall structure and active sites of PtmA2 in both the adenylation and thioesterification conformations.
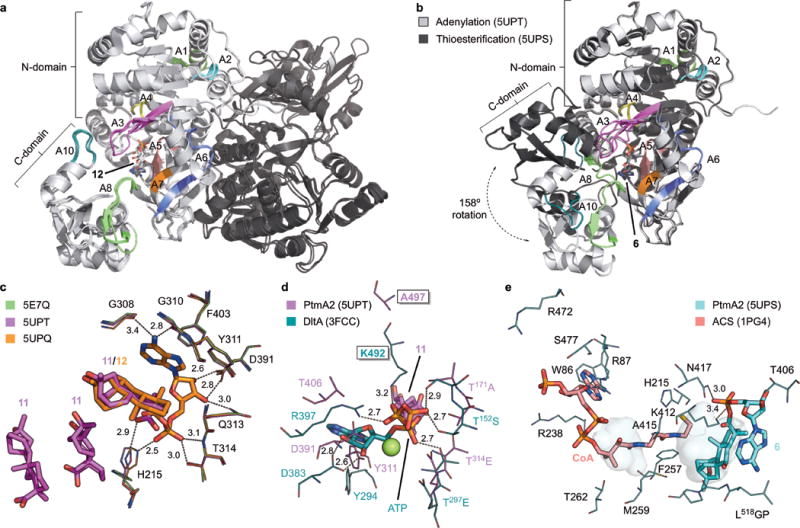
a, Superposition of the three structures of PtmA2 in the adenylation conformation (5E7Q, 5UPQ, and 5UPT). The dimer (monomers shown in light and dark gray) interface is along the length of both N-terminal domains. The core motifs are colored and labeled. Adenylate 12 is bound in the active site cavity. b, Monomers of the adenylation (5UPT, light gray) and thioesterification (5UPS, dark gray) conformations. The C-terminal domains diverge at the A8 hinge residue resulting in a 158° rotation. The core motifs are colored and labeled for the thioesterification conformation. Adenylate 6 is bound in the active site cavity. c, The adenylation active site in the apo form (5E7Q, green), in complex with three molecules of 11 (5UPT, magenta), and in complex with 12 (5UPQ, orange). d, The putative ATP binding site in PtmA2 (5UPT, magenta) overlaid with ATP, Mg2+ (green sphere), and residues from DltA (3FCC, teal)28. Ala497 (Lys492 in DltA) was proposed to be the residue that prevents adenylation from occurring. Free acid 11 is behind ATP. e, The putative CoA binding site in PtmA2 (5UPS, cyan) overlaid with CoA and interacting residues in ACS (1PG4, salmon)38. Without CoA bound, the pantetheine tunnel (shown as pockets) in PtmA2 is blocked by Phe257, Met259, and Ala415. All ligands are shown as sticks. Residues discussed in the text are depicted as lines. Dashes indicate residue–ligand interactions with distances in Å listed.
