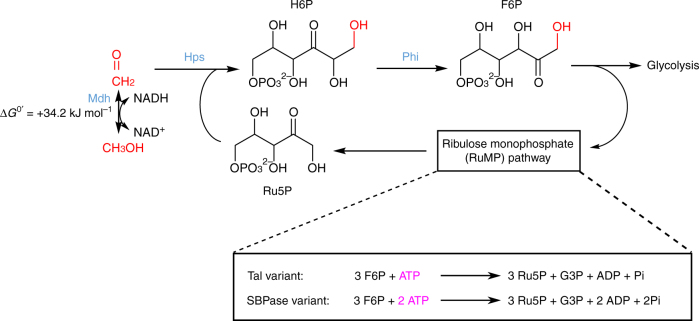
Fig. 1.
The pathway of methanol assimilation in E. coli. Heterologous enzymes required for methanol assimilation in E. coli are shown in blue. Methanol dehydrogenase (Mdh) catalyzes the thermodynamically uphill NAD-dependent oxidation of methanol to formaldehyde (ΔG0′ = +34.2 kJ mol−1), which is ligated to ribulose 5-phosphate (Ru5P) in an aldol reaction catalyzed by hexulose phosphate synthase (Hps). The resulting hexulose (H6P) is isomerized by phosphohexulose isomerase (Phi) to fructose 6-phosphate (F6P), which is metabolized through native E. coli central metabolism. A portion of the F6P must be used to regenerate Ru5P through the ribulose monophosphate (RuMP) cycle to enable further formaldehyde assimilation. Two major variants of this cycle are known, the transaldolase (Tal) and sedoheptulose bisphosphatase (SBPase), which are named for the enzyme that produces sedoheptulose 7-phosphate, and differ in their ATP requirements. The red moiety denotes the position of assimilated methanol. H6P: D-arabino-3-hexulo-6-phosphate, G3P: glyceraldehyde 3-phosphate