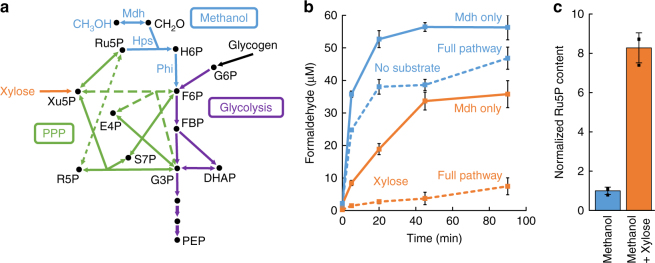
Fig. 2.
Insufficient Ru5P concentration in carbon-starved cells limits formaldehyde assimilation. a Detailed enzymatic reactions and metabolic pathways involved in methanol metabolism, showing the entry points of glycogen (the primary carbon source during starvation) through glycolysis, and xylose through the pentose phosphate pathway (PPP). Heterologous reactions unique to methanol assimilation are shown in blue. Glycolysis reactions are shown in purple, and PPP reactions in green. Dotted and dashed green lines added for clarity. b Formaldehyde levels over time after addition of 250 mM methanol to starved cells of E. coli MG1655(DE3) ΔfrmA with either no additional substrate (blue) or 6 g L−1 xylose (orange). Solid lines represent cells expressing only Mdh, and dashed lines denote cells expressing the full methanol assimilation pathway (Mdh, Hps, and Phi). c Relative ribulose 5-phosphate (Ru5P) concentration in cells with no substrate or xylose. Error bars represent s.d. of n = 3 biological replicates (three individual colonies). H6P: D-arabino-3-hexulo-6-phosphate, F6P: fructose 6-phosphate, G6P: glucose 6-phosphate, FBP: fructose 1,6-bisphosphate, DHAP: dihydroxyacetone phosphate, G3P: glyceraldehyde 3-phosphate, PEP: phosphoenolpyruvate, E4P: erythrose 4-phosphate, S7P: sedoheptulose 7-phosphate, R5P: ribose 5-phosphate, Ru5P: ribulose 5-phosphate, Xu5P: xylulose 5-phosphate