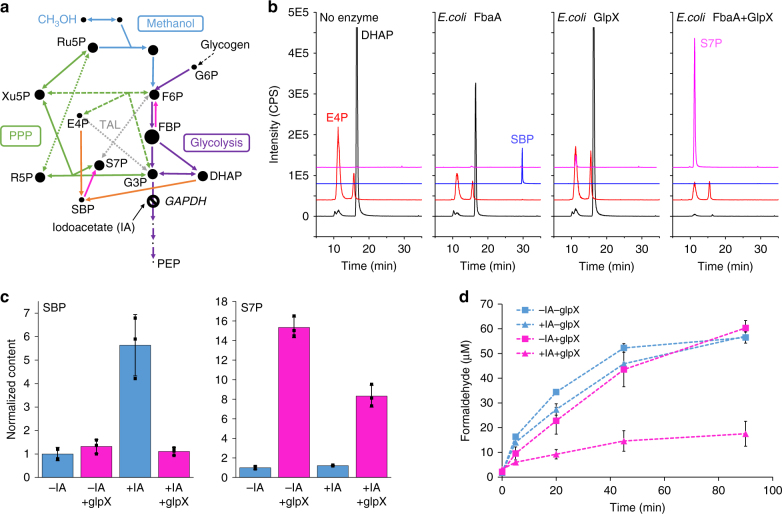
Fig. 5.
Ru5P regeneration is mediated by activation of the SBPase variant of the RuMP pathway. a Pathways involved in methanol metabolism, additionally showing the sedoheptulose bisphosphatase (SBPase) variant of the ribulose monophosphate (RuMP) pathway, where SBP is produced from DHAP and E4P by FbaA (orange line), and dephosphorylated to S7P by SBPase (pink line), compared to the transaldolase (TAL) pathway (gray line), where S7P is produced from F6P and E4P. Dot sizes indicate relative metabolite concentrations in cells treated with IA and overexpressing glpX, compared to untreated cells not overexpressing glpX. b LC-MS/MS analysis of assays with purified E. coli FbaA and GlpX, demonstrating that GlpX has SBPase activity. Black = DHAP, Red = E4P, Blue = SBP, Pink = S7P. c Relative SBP (left) and S7P (right) levels in cells treated with or without iodoacetate and expressing glpX (pink) or not (blue). d Formaldehyde timecourse in transaldolase-deficient E. coli cells (MG1655(DE3) ΔfrmA talA(K131A) ΔtalB) treated with 250 mM methanol, either expressing (pink) or not expressing (blue) glpX. Triangles denote cells treated with IA, and squares denote untreated cells. Error bars represent s.d. of n = 3 biological replicates (three individual colonies)