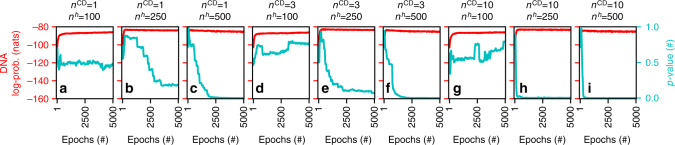
Fig. 3.
SET-RBM evolution towards a scale-free topology on the DNA dataset. We have considered three cases for the number of contrastive divergence steps, nCD = 1 (a–c), nCD = 3 (d–f), and nCD = 10 (g–i). Also, we considered three cases for the number of hidden neurons, nh = 100 (a, d, g), nh = 250 (b, e, h), and nh = 500 (c, f, i). In each panel, the x axes show the training epochs; the left y axes (red color) show the average log-probabilities computed for SET-RBMs on the test data with AIS33; and the right y axes (cyan color) show the p-values computed between the degree distribution of the hidden neurons in SET-RBM and a power-law distribution. We may observe that for models with a high enough number of hidden neurons, the SET-RBM topology always tends to become scale-free