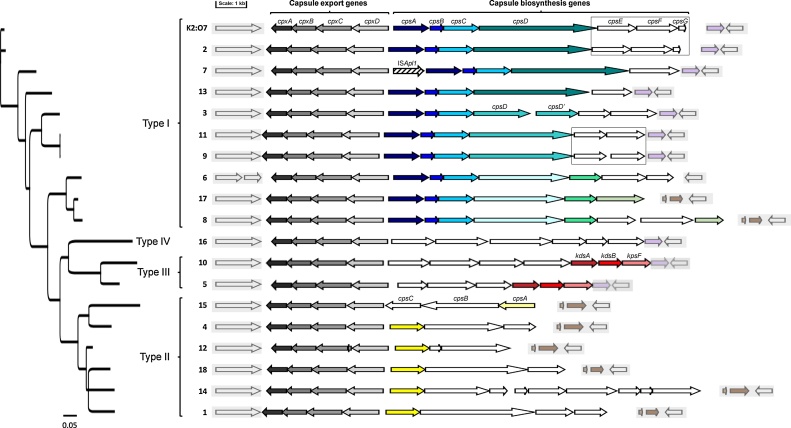
Fig. 1.
Schematic comparison of the complete capsule loci of A. pleuropneumoniae serovars 1–18 and K2:O7. The capsule loci are arranged according to phylogenetic similarity, as indicated by the tree on the left, and are clustered into their respective CPS types (I–IV) as indicated by the labeled brackets. All loci are flanked by the modF gene at the start (white arrow in shaded grey box; note an internal stop codon is present in the serovar 6 modF sequence), and ydeN at the end, preceded either the by the 552–555 bp hypothetical gene or the 114 bp hypothetical and partial lysA genes (white arrow, preceded by either mauve or brown arrows, respectively, in shaded grey box). The capsule export genes, cpxABCD, are indicated as reverse oriented arrows shaded black to light grey, respectively. The genes of the respective serovar CPS biosynthetic loci are named as follows: cps2ABCDEFG (for both K2:O7 and serovar 2); cps7ABCDE, cps13ABCDE, cps3ABCDD’EF, cps11ABCDEF, cps9ABCDEF, cps6ABCDEFG, cps17ABCDEF, cps8ABCDEFGH, cps16ABCDEF, cps10ABCD kdsAB kpsF, cps5ABC kdsAB kpsF, cps15ABC, cps4ABC, cps12ABB’, cps18ABC,cps14AB1B2B3CDEFG, cps1ABCD. The core cpsABC genes conserved in all of type I CPS loci are indicated as the dark, medium, and light blue arrows, respectively (note the extra gene at the start of the serovar 7 biosynthetic locus, shown as a striped arrow, indicates the ISApl1 insertion present in the AP76 strain, and is not part of the biosynthetic locus). The cpsD genes in the type I loci are indicated in different shades of teal, according to similarity greater than 50% identity (note in serovar 3, an internal stop codon has resulted in two orfs, cps3D and cps3D’). In serovars 6, 17 and 8, the cpsE genes share >80% identity and are shown as bright green arrows; and the last gene in the serovar 8 locus, cps8H, shares 94% identity with the C-terminal half of the serovar 17 cpsF gene, as indicated by the olive shaded arrows in the respective loci. The core genes in the type III CPS loci (kdsAB and kpsF) are indicated by the dark red, bright red, and pink arrows, respectively, at the ends of the serovar 5 and 10 loci. In the type II loci, the conserved core cpsA gene is shown as a yellow arrow. The white arrows in each biosynthetic locus indicate genes unique to each serovar. As expected, the K2:O7 CPS locus shares 96% identity across the entire sequence with that of serovar 2 (with nucleotide differences being mainly found in the cpsABC genes), and the specific cps2EFG genes found at the ends of both of these loci are boxed. Serovars 9 and 11 share 99% identity across their cpsEF genes (also shown boxed), with only a single nucleotide difference resulting in an alternate start codon for the cpsF gene in each locus. The Neighbour-Joining tree shown at the left of the figure was constructed using the Tamura-Nei algorithm with 1000 bootstraps, and the width of the line underneath it shows a 5% nucleotide difference (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).