FIGURE 1.
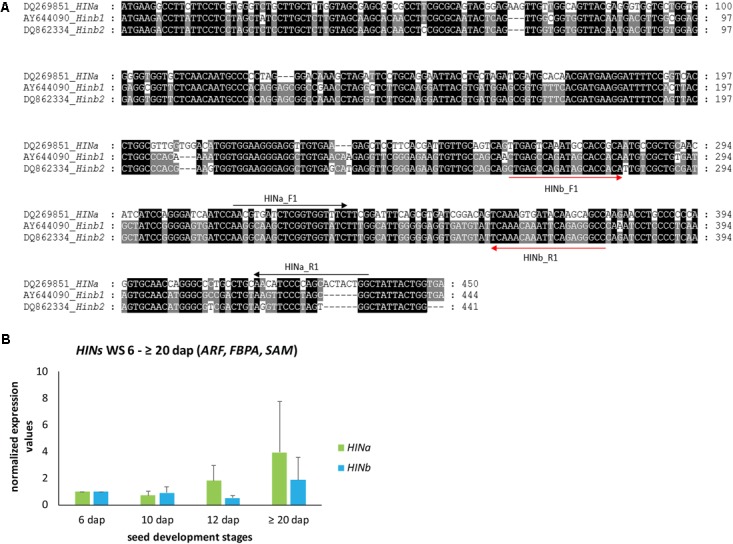
Gene alignment and quantification of HINa and HINb transcripts in WS during barley endosperm development. (A) cDNA alignment of HINa, Hinb1, and HINb2 performed by MEGA7.0.21 (Kumar et al., 2016) and visualized by GenDoc (Nicholas and Nicholas, 1997). Conserved percentage is shown as the following: black 100%, dark gray 80%. (B) Bar graph describes the average over three biological replicates of the normalized transcripts from HINa and HINb at 6, 10, 12, and ≥20 dap with the most stable RGs (ARF, FBPA, and SAM). Bars represent standard deviation. For statistical analyses, we performed a Student’s t-test. Note the trend of all HIN transcripts to increase between 6 and ≥20 dap.
